Shanghai University
Article Information
- SU Rongguo(苏荣国), CHEN Xiaona(陈小娜), WU Zhenzhen(吴珍珍), YAO Peng(姚鹏), SHI Xiaoyong(石晓勇)
- Assessment of phytoplankton class abundance using fluorescence excitation-emission matrix by parallel factor analysis and nonnegative least squares
- Chinese Journal of Oceanology and Limnology, 2015, 33(4): 878-889
- http://dx.doi.org/10.1007/s00343-015-4179-6
Article History
- Received Jul. 31, 2014
- accepted in principle Nov. 18, 2014
- accepted for publication Dec. 5, 2014
2 Tianjin University Center for Marine Environmental Ecology, School of Environmental Science and Engineering, Tianjin University, Tianjin 300072, China
Phytoplankton are the basis of food webs in marineecosystems,affecting global carbon cycles and marineecological process(Falkowski,1994; Bains et al., 2000). Phytoplankton usually concentrate off estuaries and along coastal zones where anthropogeniceutrophication affects water bodies(Søndergaard and Jeppesen, 2007). Potentially toxic algal blooms can cause significant threats to the economic and environmental value of natural waters. Investigationsof the population dynamics in phytoplanktoncommunities are important for evaluating theecological status of coastal seawater regions.
Fluorescence spectroscopy,well known as a rapid,simple and sensitive analytical technique for thediscrimination of phytoplankton taxonomic groups,has drawn increased attentions in recent years(Seppälä and Olli,2008; Proctor and Roesler, 2010; Alexander et al., 2012). Beutler et al.(2002)developed afluorometric method for the differentiation of algalpopulations in vivo and in situ using five differentexcitation wavelengths centered at 450,525,570,590 and 610 nm and acquiring the fluorescence response at680 nm,which could distinguish phytoplankton speciesinto four groups: blue algae,green algae,brown algae(diatoms and dinoflagellates), and cryptophyceae. Thistechnique is the most widely-used approach and hasbeen implemented in commercial instruments(Richardson et al., 2010; Goldman et al., 2013).
Another widely used technique is utilizingphotosynthetic and photoprotective pigmentsanalyzed by high performance liquid chromatography(HPLC)to differentiate phytoplankton classes. Theidentification of different algal groups in seawaterbased on the analysis of specific accessory pigmentshas increased in recent decades,mainly due to thedevelopment of HPLC analytical techniques(Jeffrey and Hallegraeff, 1980) and chemometric methods,such as multiple regression analysis(Gieskes and Kraay, 1983) and CHEMTAX(Mackey et al., 1996).CHEMTAX,a matrix-factorization program runningin MATLAB,uses factor analysis and a steepestdescent algorithm to determine the best fit to thepigment data matrix based on an initial pigment ratiomatrix for the algae classes. The HPLC-CHEMTAXprogram has proven to be a solid method to calculatethe abundance of phytoplankton taxonomic groups.However,pigment analysis by HPLC requires askilled professional operator, and it is time consuming and expensive,which make it ill-suited for use on alarge number of samples in a short time or for in situapplications.
Three-dimensional(3D)fluorescence,also calledexcitation-emission matrix(EEM)or EEM plots,canprovide a “fingerprint” consisting of a threedimensional(3D)emission and excitation intensityplots(Divya and Mishra, 2007). In other words,information relevant to EEMfluorescence spectroscopycan be entirely acquired by changing the excitationwavelength and the emission wavelengthsimultaneously. The various phytoplankton groupspossess variations in their accessory pigmentcompositions, and consequently,in their spectralproperties. Hence,spectral fluorescence signals can beused to differentiate the phytoplankton communitiesbased on the composition of their differing accessorypigments( Seppälä andOlli,2008; Simis et al., 2012).However,in multicomponent mixtures,thefluorescence signals are heavily overlapped,so aresolving method is required. Parallel factor analysis(PARAFAC)can successfully deconvolute the EEMfluorescence spectrum of a complex mixture into itsindividual fluorescence components. It provides aunique solution for the EEM dataset of multicomponentmixtures(Harshman,1970). The EEM fluorescencespectrum combined with PARAFAC can be used forobtaining qualitative and quantitative informationabout the multifluorophores present in the sample(Moberg et al., 2001). Fluorescence EEMs collectedfor samples give rise to three-way data,which can bearranged in a cube. The main advantage of a three-wayanalysis is that it can predict the concentration of anindividual component in the presence of any numberof uncalibrated constituents(Divya and Mishra, 2007).Over the last few decades,the combination of EEMs and PARAFAC have been successfully used to separatecomplex mixtures into their individual fluorescencecomponents(Bro,1999; Andersen and Bro, 2003; Stedmon and Markager, 2005). This technique hasbeen applied to characterize dissolved organic matter(Stedmon and Markager, 2005),polycyclic aromatichydrocarbons(Bosco and Larrechi, 2007),acetylsalicylic acid,paracetamol,caffeine(Alves and Poppi, 2009), and pigment extracts(Moberg,2001).Therefore,it is highly useful for solving analyticalproblems involving a complex matrix.
In this paper,the main purpose was to develop asimple,low-cost and sensitive fluorometric methodfor differentiating phytoplankton taxonomic groups inmarine environment by EEMs,PARAFAC and NNLS.To date,the technology has not yet been reported.
2 METHOD2.1 Cultures
Phytoplankton cultures were maintained at theculture facility of the Marine Pollution Eco-chemistryLaboratory in the Ocean University of China. Fortyone phytoplankton species,representing five divisions,were maintained as batch cultures in 250-mL conicalflasks. Each culture had two duplicates. In this study,the phytoplankton species consisted of twenty-eightmarine algae and thirteen freshwater algae,as listed inTable 1. All marine algae were grown in f/2 media at20°C,under an irradiance of 80 and 160 μmol quanta/(m2∙s)for a 12 h:12 h light:dark cycle. The culturemedium for the diatoms contained an addition ofNa2SiO3∙9H2O(Drinovec et al., 2011). The freshwateralgae were grown in BG-11 or SE medium(Clarke et al., 1987). The cell suspensions(10 mL)were filteredthrough 25-mm glass fiber filters(Whatman GF/F,0.7-μm pore size)under low vacuum(<0.3 kPa) and dim light to prevent the degradation of pigments. Afterfiltration,the filters were folded,wrapped in aluminumfoil and stored at -20°C. When analysis,the pigmentswere extracted from the filters in 10 mL extractionsolvent(N,N-dimethylformamide: DMF) and wereleft in the dark for 2 h at 4°C.
The mixtures were prepared from the exponentialgrowth phase of the five different taxonomic groups:Bacillariophyta,Chlorophyta,Dinophyta,Cryptophyta and Cyanophyta. The chlorophyll a(Chl a)concentration was determined by absorptionspectroscopy(SHIMADZU UV2550) and Jeffrey and Humphrey’s trichromatic equations. The mixtures oftwo different algal species were prepared so that theChl a of the dominant species accounted for 80% or60%.
2.3 Field samplingThe 85 surface-water samples were collected in theChangjiang River estuary in March 2013(Fig. 1). Theseawater samples,which varied in volume from 500to 1 000 mL depending on the sampling region,wereimmediately filtered through GF/F(47-nm diameter)glass-fiber filters using a vacuum pump. The filterswere stored at -20°C for 24 h under dim-lightconditions to prevent photooxidation of the pigments.When analysis,the pigments were extracted from thefilters in 10 mL extraction solvent(N,N-dimethylformamide: DMF) and were left in thedark for 2 h at 4°C. The pigment extracts from thefield samples were divided into two parts: one formeasuring the excitation-emission fluorescencespectra and the other for the HPLC-CHEMTAXanalysis.
 |
| Fig. 1 Stations for seawater samples in the Changjiang River estuary in March 2013 |
The pigment extracts were kept in dark placebefore determination. Fluorescence spectra werecollected by a Fluorolog3-11 fluorescence spectrophotometer(Jobin Yvon,France). The excitation wavelength was scanned from 350 to 700 nm at 5 nmintervals, and emission was measured from 600 nm to750 nm with a step of 5 nm.
2.5 HPLC analysisThe pigment extracts(100 μL)were automaticallydiluted to 80% with water immediately beforeinjection to improve the peak shape(Wright and Jeffrey, 1997). The extracts were then injected into aWaters Alliance 2695 HPLC system coupled with aWaters 2996 photodiode array detector and a Waters2475 Multi-λFluorescence Detector. Chromatographicseparation was performed using a C8 column forreverse-phase chromatography(150 mm×4.6 mm,3.5-μm particle size,100 Å pore size)(Zapata et al., 2000). The mobile phases were:(A)methanol:acetonitrile: aqueous pyridine solution(50:25:25,v/v/v) and (B)methanol: acetonitrile: acetone(20:60:20,v/v/v). The identification of the pigmentswas performed with retention-time data and comparison of the absorption spectra with those ofpure-pigment st and ards and a sample of mixedst and ards from known cultures. Then,the CHEMTAXsoftware was used to calculated the contribution ofthe various phytoplankton taxonomic groups to totalChl a using the concentration of the accessorypigments detected by HPLC and the pigment ratiosfor the phytoplankton species common in estuaries and coastal areas(Mackey et al., 1996).
2.6 Data processing 2.6.1 Data preprocessingAll fluorescence spectra were processed byMATLAB 6.5. Rayleigh and Raman scattering peakscould create problems for quantitative analysis and display of the EEMs. The Rayleigh and Raman scattering peaks were eliminated from the EEMs and then the missing regions were filled in using a threedimensional Delaunay interpolation of the surroundingdata points(Zepp et al., 2004),it had been proved thatthe expected interpolation error with this techniquewas within the range of measurement error for theprimary observations and the scattering correctionwas effective for individual emission scans and forthe entire EEM. All of the spectral data were thenst and ardized using the following formula:

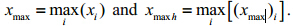
The data obtained from multiple samples werecombined into a three-way data array: 911 samples ×31 emissions × 71 excitations. We used a leverage and loading technique(Stedmon and Bro, 2008)toidentify the outliers. Seven samples were removedfrom the entire EEM dataset, and a final dataset(875samples × 31 emissions × 71 excitations)was obtainedbefore running the final PARAFAC model.
2.7 Parallel factor analysis(PARAFAC)The final dataset was modeled by the PARAFACtechnique,which has been described by Bro(1999) and Stedmon et al.(2003). The analysis was performedin MATLAB 6.5 using the “N-way toolbox forMATLAB”. PARAFAC decomposed the data signalinto a set of trilinear terms and a residual array asfollows:

It is of the utmost importance to determine thecorrect number of components for the PARAFACmodel. At present,the determination of the appropriatenumber of factors was based on split-half analysis,analysis of the residuals, and comparison of thespectral properties of each component(Christensen et al., 2005). However,in this study,the dataset is notlarge, and the compositions of fluorophores in thealgal extracts span a large range,so it is difficult tovalidate the PARAFAC model using split-half analysis(Stedmon and Bro, 2008). Therefore,the determinationof the appropriate number of factors was based on theanalysis of the residuals and an examination of thespectral properties of the individual components. Forthe analysis of the residuals,the residuals should becharacterized by small systematic variations and contain little structure. In other words,the modelshould have a low sum of the squares of the residuals.The presence of a region with a peak next to thenegative values in the residuals for many samplessuggests that the model does not fit well. This methodalone was not sufficient for the determination of thenumber of factors. Therefore,the spectral propertiesof each component must be examined. Excitationspectra can have single or multiple maxima,but theemission spectra should only exhibit a single emissionmaximum. Additionally,the excitation and theemission spectra of the fluorophores often overlapslightly.
After the PARAFAC model was validated,Bayesian discriminant analysis(BDA)(Li and Anderson-Sprecher, 2006)was used to analyze thecapability of the fluorescence components from thePARAFAC model to discriminate the phytoplanktongroups.
2.8 Assessment of the phytoplankton speciesThe data of the fluorescence components from oneduplicate culture was used as training data,the otherwas used as the test data. For the training data,hierarchical cluster analysis(HCA)was applied to allfluorescence component compositions of eachphytoplankton taxonomic group, and the referencespectra of the fluorescence components were obtained.HCA(Bona and Andrés,2007)is able to find variableswith similar properties between different samples and sample types,thus allowing us to simplify ouroperation by finding the structure or patterns in thepresence of chaotic or confusing data.
Then,the multivariable linear regression modelresolved by NNLS(Zhang et al., 2010)was used todifferentiate the phytoplankton populations based onthe reference spectra. NNLS was a constrainedversion of the least squares problem where thecoefficients are not allowed to become negative.Qualitative and relatively quantitative analyses of thesingle species(the other half/duplicate),laboratory mixtures and the field samples were performed. The regression equation is:

The estimation of the regression coefficientsspecific to the various phytoplankton taxonomicgroups was very important for the prediction of thephytoplankton community structure.
3 RESULT 3.1 PARAFAC Model validationA fifteen-component PARAFAC model wasvalidated in this study. A typical example of themeasured,modeled and residual EEM data is given inFig. 2. The fifteen-component PARAFAC modelcaptured the bulk features in the measured EEM,asindicated by the low residual(difference betweenmeasured and modeled data) and small systematicvariations that are shown in Fig. 2.
 |
| Fig. 2 Typical example of contour plot of a measured EEM and PARAFAC modeling result for a sample showingmeasured,modeled, and residual EEM |
Figure 3 shows a contour plot of each component,exhibiting rounded peaks,found by a fifteencomponent PARAFAC model. They represent fifteendifferent components that are present in the dataset;however,not all fifteen fluorescence components arenecessarily present in all measured samples. Figure 4 shows a visual inspection of the emission and theexcitation loading obtained by applying the fifteenfactor PARAFAC model. The components represent different groups offluorophores; all of thefluorescencecomponents except for component 4 had one emissionmaximum with one or more excitation maxima,as isoften seen in common material fluorophores.Consequently,the PARAFAC model of 15 factorswas used.
 |
| Fig. 3 The fifteen different fluorescent components generated from the PARAFAC model |
 |
| Fig. 4 Excitation(dotted line) and emission(solid line)spectra resulting from fitting a fifteen-component PARAFAC modelto EEM data |
Based on the 15 fluorescence components from thePARAFAC model,the five phytoplankton taxonomicgroups were correctly classified by BDA(Fig. 5).
 |
| Fig. 5 BDA of the 15 fluorescent components from the PARAFAC model
Baci: Bacillariophyta; Chlo: Chlorophyta; Dino: Dinophyta; Cyan: Cyanophyta; Cryp: Cryptophyta. |
HCA was implemented on the 15 fluorescencecomponents of the training data from the PARAFACmodel, and a database of 61 reference spectra wasestablished. It was used as criteria to discriminatephytoplankton taxonomic groups by the multivariable linear regression model resolved by NNLS. The reference spectra are shown in Fig. 6. The differencesamong the reference spectra of the five taxonomicalgal groups were significant. This reveals that eachtaxonomic phytoplankton group possesses the specificcomposition of fluorophores.
 |
| Fig. 6 The reference spectra of the fluorescent components of the five algal taxonomic groups obtained fromthe training data
RS: reference spectrum. |
When NNLS was utilized to discriminate thesamples of single algal species from different culturelights and different growth stages,the correctdiscrimination ratios(CDRs)of the five phytoplanktontaxonomic groups were 100%. The average relative contents estimated were 81.9% to 92.3%(Table 2). The relative contents of most samples were estimatedto be above 80%. The samples with relative contentsestimated to be below 80% were mainly from theearly growth stages,possibly due to their significantlylower biomass.
For mixture samples,when the dominant algaltaxonomic groups accounted for 80% of the mixtures,their CDRs ranged from 84.8% to 100.0%,with theaverage relative contents estimated from 62.5% to77.5%(Table 3). When the dominant algal taxonomicgroups accounted for 60% of the mixtures,theirCDRs ranged from 80.0% to 88.5%, and the averagerelative contents estimated were from 57.0% to74.6%. The method could also differentiate thesubdominant algal taxonomic groups in 143 of the195 samples,which indicates an average CDR of 73.3%. When the subdominant algal taxonomicgroups accounted for 40% of the mixtures,theirCDRs ranged from 72.4% to 100.0%,with the averagerelative contents estimated from 37.8% to 53.3%.When the subdominant algal taxonomic groupsaccounted for 20% of the mixtures,their CDRs rangedfrom 53.8% to 83.8%,with the average relativecontents estimated from 18.9% to 43.1%. ForBacillariophyta,Chlorophyta and Cryptophyta,theirCDRs were above 80.0% even when their relativecontents were as low as 20.0%.
Sixteen of the 85 field samples collected from theChangjiang River estuary were analyzed by bothHPLC-CHEMTAX and the developed fluorometrictechnique(Table 4). The HPLC- CHEMTAX analysisrevealed that Bacillariophyta was the dominant algalgroup in the 16 samples,accounting for 42.2% to100.0% of the total Chl a, and the subdominant algalgroups comprised Dinophyta,Chlorophyta and Cryptophyta,contributing 3.9% to 29.6% to the totalalgal abundance. The results of the developedfluorometric technique also showed that the dominantalgal groups of the 16 samples were all Bacillariophyta,which accounted for 53.1% to 99.7% of the total Chla, and the subdominant algal groups of most samplesmainly comprised Dinophyta,Chlorophyta, and Cryptophyta,contributing 9.7% to 46.9% to the totalalgal abundance. Only several samples were predictedto have Cyanophyta as one of the subdominant algalgroups(Table 4). The differentiation results by thefluorometric technique were in good agreement withthose of HPLC-CHEMTAX.
 |
85 field samples were analyzed by the fluorometrictechnique developed. Bacillariophyta was thedominant algal taxonomic group in all samples,accounting for 53.1% to 100.0% of the total algalabundance. The results also indicated that Dinophytawas the subdominant algal group at stations 42,43,44,54,59,63,66,67 and 71,with 0.3% to 46.9% ofthe total algal abundance. For stations 23,29,56,57,58 and 64 near the Changjiang River mouth and theHangzhou Bay,Cryptophyta was the subdominantalgal group,which accounted for 3.4% to 12.8% ofthe total algal abundance. Cyanophyta was also foundto be the subdominant algal group at stations 1,7 and 13 near the boundary between the East China Sea and the Yellow Sea,which accounted for 25.3% to 34.2%of the total algal abundance. The subdominant algalgroup for the remaining stations was Chlorophyta,contributing 0.1% to 23.7% of the total algalabundance. In conclusion,Bacillariophyta was the dominant algal taxonomic group in the survey area, and Dinophyta,Chlorophyta,Cryptophyta orCyanophyta was the subdominant contributor at moststations. These results were in accordance with thoseof Kong in the same sea region(Kong,2012); Kongfound that Bacillariophyta was the dominantphytoplankton class that contributed the most to Chl ain most cases. The subdominant phytoplankton classwas Dinophyta,Cryptophyta,Chlorophyta orCyanophyta. Bacillariophyta,Dinophyta,Cryptophyta and Chlorophyta were always distributed in the upperlayer of the stratified water in the flume,whereasCyanophyta occurred mainly outside the plume.
4 DISCUSSION 4.1 Number of components identifiedThe absence of large peaks in the residual EEMs(Fig. 2)shows that the fifteen-component PARAFACmodel can explain the majority of the measuredfluorescence. However,several small peaks near thefirst-order Rayleigh scattering trough could not bemodeled. The small peaks may also indicate thepresence of weakly fluorescing components due toinstrument noise,inner filter or scattering effects,which were the fractions with such a low fluorescencethat the PARAFAC model was unable to resolve them.The ranges of the sums of the squared residuals forthe fifteen-component model are 0.002 9–0.007 6,0.002 7–0.007 4,0.002 7–0.007 6,0.003 6–0.007 4 and 0.005 0–0.006 5 for Bacillariophyta,Chlorophyta,Dinophyta,Cyanophyta, and Cryptophyta,respectively. The averages of the sums of the squaredresiduals for the five divisions are 0.004 8,0.005 1,0.004 5,0.005 3 and 0.006 1,respectively. The valuesare all below 1% relative to the highest fluorescenceintensity. It is clear that the presence of the sum of thesquared errors for the fifteen-component PARAFACmodel indicates that the feature regions of the EEMsin the dataset were well described by the PARAFACmodel.
4.2 Analysis of component characteristicsVisual analysis of the components identified(Fig. 3)reveals that it is unlikely that each componentrepresents a specific fluorophore; some componentsrepresent a group of fluorophores with very similarfluorescence characteristics and variability. Forcomponent 4,the existence of multiple emissionb and s within a component might imply the presenceof different fluorophores that were chemically boundtogether. Components 6 and 9 exhibit emission atlonger wavelengths than the other components,whichsuggests that they might contain more conjugated fluorescence molecules than the other components(Sharma and Schulman, 1999). The results suggestedthat the model was successful at grouping thefluorophores present into groups with similarfluorescence/molecular structures, and the spectraretrieved were representative of the “pure” analytes inthe samples.
4.3 Model predictionsThe phytoplankton biomass in each mixture samplewas predicted using the developed fluorometrictechnique. The relationships between the observed and the predicted phytoplankton biomass for theNNLS model are shown in Fig. 7, and they were nearlylinear. The regression coefficients obtained from theNNLS model can be compared with the measuredones. For Bacillariophyta,the predictions by NNLShad a small amount of scatter, and the predictions forthe observed biomass from 0.2 to 0.5 were biasedslightly high. For Dinophyta,the predicted biomasshad some scatter and a small amount of bias. Thescatter in the prediction of Chlorophyta,Cryptophyta and Cyanophyta was smaller, and the predictions hada slightly low bias with this method.
 |
| Fig. 7 Relationship between the observed and the predicted phytoplankton biomass for the NNLS model |
In this paper,algal pigment extracts were studiedusing fluorescence EEMs in combination withPARAFAC and the NNLS method to develop a fluorescence differentiation technique forphytoplankton populations. The fluorometrictechnique developed succeeded in the qualitative and quantitative discrimination of five algal taxonomicgroups: Bacillariophyta,Chlorophyta,Dinophyta,Cryptophyta, and Cyanophyta. For the samplescollected from the Changjiang River estuary,theresults from the fluorometric method were in goodagreement with those from HPLC-CHEMTAX. Thiswork demonstrated that the developed fluorometrictechnique is a powerful tool for quickly analyzing alarge number of algal samples in situ.
| Alexander R, Gikuma-Njuru P, Imberger J. 2012. Identifying spatial structure in phytoplankton communities using multi-wavelength fluorescence spectral data and principal component analysis. Limnology and Oceanography : Methods, 10 (6): 402-415. |
| Alves J C L, Poppi R J. 2009. Simultaneous determination of acetylsalicylic acid, paracetamol and caffeine using solidphase molecular fluorescence and parallel factor analysis. Analytica Chimica Acta, 642 (1-2): 212-216. |
| Andersen C M, Bro R. 2003. Practical aspects of PARAFAC modeling of fluorescence excitation-emission data. Journal of Chemometrics, 17 (4): 200-215. |
| Bains S, Norris R D, Corfield R M, Faul K L. 2000. Termination of global warmth at the Palaeocene/Eocene boundary through productivity feedback. Nature, 407 (6801): 171- 174. |
| Beutler M, Wiltshire K H, Meyer B, Moldaenke C, Lüring C, Meyerhöfer M, Hansen U-P, Dau H. 2002. A fluorometric method for the differentiation of algal populations in vivo and in situ. Photosynthesis Research, 72 (1): 39-53. |
| Bona M T, Andrés J M. 2007. Coal analysis by diffuse reflectance near-infrared spectroscopy: hierarchical cluster and linear discriminant analysis. Talanta, 72 (4): 1 423-1 431. |
| Bosco M V, Larrechi M S. 2007. PARAFAC and MCR-ALS applied to the quantitative monitoring of the photodegradation process of polycyclic aromatic hydrocarbons using three-dimensional excitation emission fluorescent spectra: comparative results with HPLC. Talanta, 71 (4): 1 703-1 709. |
| Bro R. 1999. Exploratory study of sugar production using fluorescence spectroscopy and multi-way analysis. Chemometrics and Intelligent Laboratory Systems, 46 (2): 133-147. |
| Christensen J H, Hansen A B, Mortensen J, Andersen O. 2005. Characterization and matching of oil samples using fluorescence spectroscopy and parallel factor analysis. Anal ytical Chem istry, 77 (7): 2 210-2 217. |
| Clarke S E, Stuart J, Sanders-Loehr J. 1987. Induction of siderophore activity in Anabaena spp. and its moderation of copper toxicity. Appl ied and Environ mental Microbiol ogy, 53 (5): 917-922. |
| Divya O, Mishra A K. 2007. Multivariate methods on the excitation emission matrix fluorescence spectroscopic data of diesel-kerosene mixtures: a comparative study. Analytica Chimica Acta, 592 (1): 82-90. |
| Drinovec L, Flander-Putrle V, Knez M, Beran A, Berden-Zrimec M. 2011. Discrimination of marine algal taxonomic groups using delayed fluorescence spectroscopy. Environmental and Experimental Botany, 73 : 42-48. |
| Falkowski P G. 1994. The role of phytoplankton photosynthesis in global biogeochemical cycles. Photosynthesis Research, 39 (3): 235-258. |
| Gieskes W W C, Kraay G W. 1983. Dominance of Cryptophyceae during the phytoplankton spring bloom in the central North Sea detected by HPLC analysis of pigments. Marine Biology, 75 (2-3): 179-185. |
| Goldman E A, Smith E M, Richardson T L. 2013. Estimation of chromophoric dissolved organic matter (CDOM) and photosynthetic activity of estuarine phytoplankton using a multiple-fixed-wavelength spectral fluorometer. Water Research, 47 (4): 1 616-1 630. |
| Harshman R A. 1970. Foundations of the PARAFAC procedure: models and conditions for an “explanatory” multi-modal factor analysis. UCLA Working Papers in Phonetics, 16 (1): 1-84. |
| Jeffrey S W, Hallegraeff G M. 1980. Studies of phytoplankton species and photosynthetic pigments in a warm core eddy of the East Australian Current. I. Summer populations. Marine Ecology Progress Series, 3 : 285-294. |
| Kong F Z. 2012. Size-fraction Structure, Species Component and Pigments Analyses of Phytoplankton in the Bloom Zone Near Changjiang Estuary. PhD dissertation. Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China. (in Chinese) |
| Li Y M, Anderson-Sprecher R. 2006. Facies identification from well logs: a comparison of discriminant analysis and naïve Bayes classifier. Journal of Petroleum Science and Engineering, 53 (3-4): 149-157. |
| Mackey M D, Mackey D J, Higgins H W, Wright S W. 1996. CHEMTAX- a program for estimating class abundances from chemical markers: application to HPLC measurements of phytoplankton. Marine Ecology Progress Series, 144 : 265-283. |
| Moberg L, Robertsson G, Karlberg B. 2001. Spectrofluorimetric determination of chlorophylls and pheopigments using parallel factor analysis. Talanta, 54 (1): 161-170. |
| Proctor C W, Roesler C S. 2010. New insights on obtaining phytoplankton concentration and composition from in situ multispectral Chlorophyll fluorescence. Limnology and Oceanography : Methods, 8 (12): 695-708. |
| Richardson T L, Lawrenz E, Pinckney J L, Guajardo R C, Walker E A, Paerl H W, MacIntyre H L. 2010. Spectral fluorometric characterization of phytoplankton community composition using the Algae Online Analyser. Water Research, 44 (8): 2 461-2 472. |
| Seppälä J, Olli K. 2008. Multivariate analysis of phytoplankton spectral in vivo fluorescence: estimation of phytoplankton biomass during a mesocosm study in the Baltic Sea. Marine Ecology Progress Series, 370 : 69-85. |
| Sharma A, Schulman S G. 1999. Introduction to Fluorescence Spectroscopy. Wiley, New York. |
| Simis S G H, Huot Y, Babin M, Seppälä J, Metsamaa L. 2012. Optimization of variable fluorescence measurements of phytoplankton communities with cyanobacteria. Photosynthesis Research, 112 (1): 13-30. |
| Søndergaard M, Jeppesen E. 2007. Anthropogenic impacts on lake and stream ecosystems, and approaches to restoration. Journal of Applied Ecology, 44 (6): 1 089-1 094. |
| Stedmon C A, Bro R. 2008. Characterizing dissolved organic matter fluorescence with parallel factor analysis: a tutorial. Limnology and Oceanography : Methods, 6 (11): 572-579. |
| Stedmon C A, Markager S, Bro R. 2003. Tracing dissolved organic matter in aquatic environments using a new approach to fluorescence spectroscopy. Marine Chemistry, 82 (3-4): 239-254. |
| Stedmon C A, Markager S. 2005. Resolving the variability in dissolved organic matter fluorescence in a temperate estuary and its catchment using PARAFAC analysis. Limnology and Oceanography, 50 (2): 686-697. |
| Wright S W, Jeffrey S W. 1997. High resolution HPLC system for chlorophylls and carotenoids of marine plankton. In : Jeffrey S W, Mantoura R F C, Wright S W eds. Phytoplankton Pigments in Oceanography: Guidelines to Modern Methods. UNESCO Public, Paris, ISBN: 92-3- 103275-5, p.327-341. |
| Zapata M, Rodríguez F, Garrido J L. 2000. Separation of chlorophylls and carotenoids from marine phytoplankton: a new HPLC method using a reversed phase C8 column and pyridine-containing mobile phases. Marine Ecology Progress Series, 195 : 29-45. |
| Zepp R G, Sheldon W M, Moran M A. 2004. Dissolved organic fluorophores in southeastern US coastal waters: correction method for eliminating Rayleigh and Raman scattering peaks in excitation-emission matrices. Marine Chemistry, 89 (1-4): 15-36. |
| Zhang F, Su R G, He J F, Cai M H, Luo W, Wang X L. 2010. Identifying phytoplankton in seawater based on discrete excitation-emission fluorescence spectra. J ournal of Phycol ogy, 46 (2): 403-411. |
 2015, Vol. 33
2015, Vol. 33





