Institute of Oceanology, Chinese Academy of Sciences
Article Information
- WU Xiangwei, LIU Xiande, YU Ziniu
- Analysis of novel immune-related genes and microsatellite markers in the transcriptome of Paphia undulata
- Journal of Oceanology and Limnology, 37(4): 1301-1316
- http://dx.doi.org/10.1007/s00343-019-8154-5
Article History
- Received May. 21, 2018
- accepted in principle Jul. 11, 2018
- accepted for publication Aug. 29, 2018
2 Animal Science and Technology College, Yunnan Agricultural University, Kunming 650201, China;
3 Key Laboratory of Healthy Mariculture for the East China Sea, Ministry of Agriculture of China; Fisheries College, Jimei University, Xiamen 361021, China;
4 University of Chinese Academy of Sciences, Beijing 100049, China
Invertebrates rely mainly on innate immunity for host defense (Loker et al., 2004). This defense system is essential for the survival of invertebrates. They have developed specific modalities to detect and respond to microbial surface antigens, such as lipopolysaccharides, peptidoglycan, (1→3) β-D-glucans, and lipoproteins, as well as enzyme cascades that work together in different biological host defense systems, including complement system and antimicroorganism system mediated by toll-like receptors and peptidoglycan binding protein (Underhill and Ozinsky, 2002; Iwanaga and Lee, 2005). Generally, shellfish species have similar conserved immune components and systems to the invertebrate model animals in which immunity has been well study and characterized, such as Drosophila melanogaster and Caenorhabditis elegans (Iwanaga and Lee, 2005). However, only a small number of investigations into immunity in shellfish have been reported (Gueguen et al., 2003; Pallavicini et al., 2008; Bettencourt et al., 2010; Feng et al., 2010; Venier et al., 2011; Moreira et al., 2012).
Due to its constant contact with the external environment, the foot is one of the main targets that attack to pathogens in clams. Parasite infections are a major problem in clams and can impede the functionality of the foot, such as by reducing the ability to burrow into sediment and by limiting hemolymph movement and muscles contraction of the clam's foot (Thomas et al., 1998). The foot is also the only site where echinostome parasites accumulate in clams (O'Connell-Milne et al., 2016), which causes tissue damage and stress to the clam, results in immunosuppression, and increases vulnerability to hypoxic conditions (Jensen et al., 1999; Paul-Pont et al., 2010). Interestingly, clams have the ability to regenerate foot tissue after physical damage or parasitic penetration (Mouritsen and Poulin, 2003). Meanwhile, the stress of wound healing can be in response to immune reaction in order to expel harmful substances from the clam tissue (Lauckner, 1983).
The venerid clam Paphia undulata is a common soft-sediment bivalve species. It mainly distributes in the coastal waters of southern China and Southeast Asia and is an important shellfish resource (Leethochavalit et al., 2004). However, exogenous factors such as viral, bacterial and parasitic infections and variations in water conditions have become major threats to the clam, especially in aquaculture production (Beaz-Hidalgo et al., 2010). To date, little sequence information is available on the expressed immune genes in P. undulata. The lack of genomic resources, coupled with poor understanding of the molecular and biochemical processes, have hindered advances in disease control and aquaculture productivity for P. undulata. RNA sequencing (RNA-Seq) technology is a powerful high-throughput method for exploring trait-related transcriptome sequences (Feldmeyer et al., 2011; Shi et al., 2013). RNA-Seq has been employed for gene discovery, molecular resource development, and understanding the biological processes of specific phenotypes in shellfish species (Clark et al., 2010; Wang et al., 2011; Huan et al., 2012; Niu et al., 2013a). In this study, we used the Illumina Hiseq 2000 platform to characterize the transcriptome of foot tissue to identify the stress-response and immune-related genes in P. undulata, which will be useful for further immunity and genomic studies.
2 MATERIAL AND METHOD 2.1 Sample collection and preparationPaphia undulata specimens, with an average shell length of 44 mm, were collected from the major P. undulata farming area of the China Eastern Sea in Dongshan Island, Fujian Province, China. The foot tissue was immediately sampled and stored in liquid nitrogen, and then transferred to -80 ℃ for long-term preservation.
2.2 cDNA library preparation and Illumina sequencing for transcriptome analysisThe foot tissue from 6 clams was dissected for total RNA extraction using TRIzol (Invitrogen, USA) according to the manufacturer's instructions. Equal quantities of high-quality RNA from each clam were pooled for cDNA synthesis and RNA-seq library construction by an mRNA-seq sample preparation Kit (Illumina Inc., San Diego, CA). The cDNA library was sequenced using the Illumina Hiseq 2000 platform in 100-pair-ended mode (Biomarker Technologies).
2.3 Quality control and de novo transcriptome assemblyThe adaptor and ambiguous sequences were removed from the raw data before quality filtering and assembly. The quality screening was performed using the FastX-toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html). The reads with Q-value ≤20 were defined as low quantity sequences and were discarded from the raw data. Then de novo transcriptome assembly was carried out using Trinity (Version r20130225) (Haas et al., 2013). The clean reads were assembled into transcripts through pair-end joining and gap filling and then were clustered to be unigenes.
2.4 Sequences annotationAll unigenes were then searched for homologous sequences against public databases including Nr, Nt, and Swiss-Prot using BLASTX (version 2.2.14) (Altschul et al., 1997) with an E-value cut-off of 1e-5. The best aligning results were chosen to annotate unigenes. Blast2Go was employed to assign Gene Ontology (GO) terms to unigenes (Conesa et al., 2005). The unigenes were also aligned to the Cluster of Orthologous Groups (COG) database to predict and classify possible functions (Tatusov et al., 2000). Meanwhile, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database was also used to annotate the unigenes using an online server (http://www.genome.jp/kegg/kass/), with an E-value threshold of 1e-5.
2.5 Microsatellite marker detection and polymorphism validationMicrosatellite markers (SSRs) were screened from all unigenes using MISA software (http://pgrc.ipk-gatersleben.de/misa/misa.html). The minimum contiguous repeat units were set as dimer-6, trimer-5, tetramer-5, pentamer-5, and hexamer-5. SSR primers were obtained from the flanking regions of microsatellite locus using the software Primers 5.0 (http://www.bbioo.com/download/58-166-1.html). Thirty wild P. undulata individuals were sampled from Beihai, Guangxi Province, China and were used for the polymorphism validation of 16 SSRs. The genomic DNA was obtained from the foot tissue for each individual using the standard phenol-chloroform protocol (Sambrook et al., 1989). PCR was performed on a gradient thermal cycler (Bio-Rad, USA) with the following protocol: denaturation at 94 ℃ for 5 min; 30 cycles of 20 s at 94 ℃, 20 s at annealing temperature, and 20 min of elongation at 72 ℃; and a final elongation of 8 min at 72 ℃. PCR products were used for electrophoresis on 6% polyacrylamide gels with silver staining. The 100 bp DNA marker (GeneStar, Beijing, China) was used to determine the allele size. The software PopGen32 (1.32 version) was used to estimate the allele frequency, observed heterozygosity (Ho) and expected heterozygosity (He) (Yeh et al., 2000), and polymorphism information content (PIC) was calculated using the method described by Botstein et al. (1995).
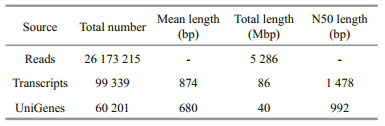
3 RESULT 3.1 Sequencing and de novo assemblyThe length distributions of transcripts and unigenes are in Fig. 1. There were 24 396 transcripts longer than 1 kb and 9 928 transcripts longer than 2 kb. All transcripts were clustered into 60 201 unigenes, of which 23 296 (38.7%) unigenes were longer than 0.5 kb and 10 434 (17.33%) were longer than 1 kb (Table 1).

|
| Fig.1 Overview of the distribution of transcripts and unigenes in the transcriptome |
|
In total, 22 260 unigenes (36.98%) were successfully annotated by public databases listed in Table 2. Among the annotated unigenes, 15 017 were assigned to one or more GO terms and were further divided into 63 functional terms (Table S1). For the cellular component category, the terms "membrane", "membrane part", and "membrane-enclosed lumen" accounted for 10.29%, 5.81%, and 2.99% of the total, respectively (Fig. 2). Whilst, in the molecular function category, a large number of unigenes were defined as "binding" (47.16%), "catalytic activity" (28.99%), and "transporter activity" (5.32%) functions. The top 5 GO terms in the biological process category were "cellular process" (14.35%), "metabolic process" (11.84%), "biological regulation" (10.71%), "response to stimulus" (8.63%), and "developmental process" (8.15%).

|
| Fig.2 Functional annotation of assembled unigenes based on the GO terms |
In total, 7 832 unigenes could be assigned to the COG classifications. The group "general function prediction" (26.59%) represented the largest cluster, followed by "replication, recombination and repair" (9.86%), "signal transduction mechanisms" (7.39%), and "transcription" (6.94%). Only 5 and 17 unigenes were assigned to the COG of "cell motility" and "nuclear structure", respectively (Fig. 3).

|
| Fig.3 Clusters of orthologous groups (COG) classification |
A total of 6 718 unigenes were predicted in 19 KEGG pathways. The top 5 metabolic pathways were "ubiquitin-mediated proteolysis" (ko04120), "purine metabolism" (ko00230), "protein processing in endoplasmic reticulum" (ko04141), "lysosome" (ko04142), and "RNA transport" (ko03013) (Table S2).
3.3 Stress-response and immune-related unigenes in the high abundant expressed sequencesThe 50 most expressed unigenes with a large number of reads were identified from the transcriptome data. BLAST sequence similarity searches indicates that these unigenes belonged to diverse functional classes, reflecting the many functions of the foot tissue, including response to stress, immune reaction, growth and complex contractile (Table 3). The classes of unigenes included the heat shock protein 70, cold shock domain protein, and Opine dehydrogenase. Moreover, other unigenes were response to immune, such as ferritin, ubiquitin carboxyl-terminal hydrolase L5, translationally controlled tumor protein, and cathepsin L (Table 3).
A total of 566 unigenes showed significant similarities to genes known to be involved in the immune system. These unigenes include pattern recognition receptors (PRRs), signal transduction, and immune effectors according to the process of immune response (Table 4).

|
A total of 4 270 SSRs were identified from 60 201 unigenes, indicating that 7.1% of the unigene sequences contained SSR loci. Detailed analysis showed that the types of SSR markers were 35.2% di-nucleotide, 52.3% tri-nucleotide, 8.9% tetra-nucleotide, 3.5% penta-nucleotide, and 0.2% hexa-nucleotide (Fig. 4). For the 16 SSRs that possessed high numbers of core sequence repeats, allele frequency analysis showed that the number of effective alleles ranged from 4 to 11, with an average of 6.4 per locus, and their PIC values varied from 0.405 to 0.890, averaging 0.705 per locus (Table 5).

|
| Fig.4 Frequency of microsatellite markers (SSRs) in term of motifs screened from the P. undulata transcriptome |

|
A total of 261 237 single nucleotide polymorphisms (SNPs) were identified in the present study, of which 40 488 were putative transitions (Ts), 220 353 were putative transversions (Tv), giving the mean Ts:Tv ratio of 1:5.44 across the transcriptome of P. undulata (Fig. 5). The A/T, G/T, and A/C SNP types were the most common. In contrast, the A/G, C/T, and C/G were the smallest SNP types. Moreover, the numbers of the two SNP types of transitions were almost identical.

|
| Fig.5 Distribution of putative single nucleotide polymorphisms (SNPs) in the P. undulata transcriptome |
Several clam species including P. undulata live in intertidal regions of the sea. The various environmental stresses such as high temperature, hypersalinity, exposure to air, and clearing organism invasion are thought to make the clam more vulnerable to body damage due to the excessive and accelerated production of harmful metabolic products, such as oxidized proteins, reactive oxygen species (ROS), and polyamine (Aguirre et al., 2005). However, clams have necessarily developed the capacity to cope with such constant stressors (Philipp et al., 2005). Our results strongly support the hypothesis, because numbers of unigenes are response to environmental stresses and immune reactions and abundantly express. Those unigenes can be classed as four groups. One group includes heat shock protein 70, cold shock domain protein, and opine dehydrogenases, which are the conserved nucleic acid-binding proteins with a role in response to heat, cold, and hypoxia. The second group includes complement component C3 and ferritin, which are generally regarded as cytokine as part of the complement pathway and the iron-independent nuclear factor kappa B pathway. The third group includes cathepsin L and ubiquitin carboxyl-terminal hydrolase L5, which are proteases involved in intracellular protein degradation. The fourth group is translationally controlled tumor protein, which possesses the capacity to bind pathogens.
Thermal stress can cause problems with folding proteins (Brun et al., 2008). P. undulata has been reported to express the constitutive form of heat shock protein 70 (Wu et al., 2014), possibly in order to make protein folding more efficient at high temperatures. Additionally, cold shock domain protein (CSDP) is an RNA chaperone and can bind to RNA to prevent the termination of transcription at low temperatures (Thieringer et al., 1998). Indeed, studies on Escherichia coli and the Zhikong scallop Chlamys farreri have shown that CSDP expressed in response to cold shock (Jiang et al., 1997; Yang et al., 2012). In the present study, the CSDP sequence of P. undulata (PuCSDP) consists of a cold shock domain (CSD) with two consensus RNA-binding motifs (Fig.S1) (Weber et al., 2002). Multiple alignment analysis also showed that PuCSDP is 52% identical to both the Haliotis diversicolor Y-Box binding protein 1 and the Aplysia californica Y-box factor. Thus, there is a possibility that CSDP can be an RNA chaperone in response to low temperature in P. undulata. Opine dehydrogenases (OpDHs) are a set of the pyruvate oxidoreductases. Studies on OpDHs in invertebrates have shown that OpDHs play an important physiological role in regulating the cytoplasmic redox balance in hypoxic conditions (Grieshaber et al., 1994). Sequence alignment showed that P. undulata OpDH (PuOpDH) had the highest identity of 54.1% with the octopine dehydrogenase of Mytilus galloprovincialis and the alanopine dehydrogenase of Crassostrea gigas (Fig.S2), which suggests a new homology of OpDH in P. undulata. The abundant occurrence of PuOpDH indicates that foots are exposed frequently to low oxygen in P. undulata.
Complement component C3 is a central component of the innate immune system and is able to detect and clear potential pathogens in association with other complement proteins (Delanghe et al., 2014). C3 expressed in high concentrations in the serum and liver of the razor clam Sinonovacula constricta (Peng et al., 2016). This study confirms that C3 is abundantly expressed in the foot tissue (PuC3), suggesting that C3 also plays an important role in the innate immune response of P. undulata. Sequence alignment showed that PuC3 was 75% identical to the known C3 sequence of Ruditapes decussatus (Fig.S3). Ferritin plays an important role in the immune response, as it can capture circulating iron to defend against infection and functions as a cytokine through the iron-independent nuclear factor kappa B pathway (Ruddell et al., 2009). Multiple alignments showed that it is most similar (95.3%) to the ferritin of R. philippinarum (Fig.S4).
In invertebrate species, translationally controlled tumor protein (TCTP) is an anti-bacterial or anti-viral factor (Cronin et al., 2009). Multiple alignment indicated that P. undulata TCTP is 61.6% identical to R. philippinarum TCTP (Fig.S5), and its high levels of expression in the foot tissue suggest it has important roles in innate immunity in P. undulata. Ubiquitin carboxyl-terminal hydrolase L5 (UCHL5) can suppress protein degradation through disassembling polyubiquitin and play an important role in the defense against pathogens (Yeh and Klesius, 2010). So far, the public database includes only one homolog of UCHL5, that of C. gigas. The UCHL5 of P.undulata has 11.1% identical to that of C. gigas (Fig. S6). Cathepsin L (CtsL) is a lysosomal cysteine protease and plays a role as chemical barrier defending against microbial invasion. Four types of CtsL (L1–L4) have been identified in bivalve species (Niu et al., 2013b). Phylogenetic analysis indicated that P. undulata CtsL is 92% identical to the CtsL3 of S. constricta, which may suggest that the CtsL of P.undulata is a new homology of CtsL3 (Figs.S7, S8).
There are a number of other highly abundantly expressed unigenes, which are not specifically involved in immune reactions but could be involved in defense mechanisms and signal transduction in immune reaction pathways, including smoothelin like protein 1, signal transducing adapter molecule 2, janus kinase and microtubule interacting protein 1-like, and cAMP-responsive element binding protein. In P. undulata, the foot tissue is more sensitive to changes in external environmental factors than other tissues, as it is in constant contact with the external environment and thus has adapted to receive external signals sensitively. The genes that abundantly expressed in the foot tissue may be involved in the pathway of signal reception and transduction.
4.2 Putative immune-related genesThe initial step of the immune response is commonly immune recognition and involves the identification of an exogenous substance, mainly via pattern recognition receptors (PRRs). Several PRRs homologies were identified in this study (Tables 4 and S3), in which lectins and toll-like receptors (TLRs) were highly expressed. Lectins are a superfamily of proteins that play crucial roles in exogenous substance recognition, agglutination, opsonization, and phagocytosis in innate immunity (Matsubara et al., 2006). Several lectins such as c-type lectin, galectin, lactose-lectin, mannose-lectin or sialic acid-binding lectin were identified from bivalve species (Kim et al., 2008; Wang et al., 2009; Song et al., 2010). In the present study, 4, 4 and 2 unigenes are homologous to C-type lectin, C-type lectin domain family, and C-type lectin domain-containing protein, respectively, all of which are calcium-dependent carbohydrate-binding lectins (van de Wetering et al., 2004). Collectins and mannan-binding lectins are also included in the family and were identified in the present study. Three unigenes each are homologous to galectin and tandem repeat galectin, which are ubiquitous in multicellular organisms and bind to β-galactosides (Janeway and Medzhitov, 2002). Two unigenes homologous to techylectin were found in the present study. Techylectin is a novel type of lectin and can recognize and bind to the exposed polysaccharides on the surface of microorganisms, to aid in microbial clearance (Bahia et al., 2010). Homologous unigenes were also found for other members of the lectin family, including fucolectin, D-galactoside-lectin, lactose-lectin, and sialic acid-binding lectins. The highly abundant occurrence of lectins in the foot tissue suggests that lectins are strongly involved in immune responses in P. undulata. Toll-like receptors initiate immune responses by recognizing a variety of PAMPs (Iwasaki and Medzhitov, 2004). Although TLRs are widely distributed in nearly all animal phyla, the research into TLRs is still quite scarce in bivalves (Song et al., 2011). In the present study, 41 unigenes are considered TLR 1, 2, 3, 4, 5, 6, 7, and 13. Their abundant expression may indicate that they play special roles in the innate immune response in the foot tissue.
After recognition of the invasive pathogens, signaling pathways are triggered. In the present study, genes involved in several of these conserved signaling pathways have been identified, including the complement pathway, TLR signaling pathway, mitogen-activated protein kinase (MAPK) signaling pathway, NF-κB signaling pathway, and apoptosis pathway, reinforcing the suggestion that multiple signaling pathways could be present in P. undulata (Table 4 and Table S4). The complement pathway is considered a crucial component in innate immunity, as it plays an important role in the innate defense against common pathogens. The pathway is a relatively complex network that comprises more than 30 serum proteins and cell surface receptors, all of which participate in a range of functions from direct cell lysis to the enhancement of B and T cell responses (Carroll, 2004). Although the complement pathway has not been extensively studied in bivalve species, several sequences with similarity to complement pathway components such as C1q domain, C3, and B factor-like proteins have been found in bivalve species (Gourdine and Smith-Ravin, 2007; Zhang et al., 2008; Gerdol et al., 2011). Our results demonstrated that the complement pathway in the P. undulata foot tissue was almost complete compared to the KEGG reference pathway (ko04610) and a previous study on the complement pathway (Dunkelberger and Song, 2010). However, some complement components were absence from the transcriptome of P. undulata, which may be due to a distinction between the immune systems of bivalve species and vertebrate species. In addition, the low sequencing coverage for the present study may be partially responsible for the absence of some complement components. The TLR signaling pathway is consist of five successive parts: receptors, adaptors, kinases, transcription factors, and effectors in mammals and invertebrate species (Jeong and Lee, 2011). Each part also comprises several different members, depending on the type of TLRs. The TLR signaling pathway discovered here was almost complete compared to the KEGG reference (ko04620) and previous reports about TLRs (Akira, 2003; Takeda and Akira, 2005).
The elimination of pathogens and the management of environmental stress through innate immunity are directly reliant on the production of immune effectors. Several classic immune effectors were identified in the present study, including heat shock proteins, antioxidant enzymes, lysozymes, and mucins. They were abundantly expressed in the P. undulata foot tissue (Tables 4 and S5), probably indicating widespread participation in the innate immunity occurring in the foot tissue. HSPs play important roles in an NF-κB signaling pathway and apoptosis pathway (Parcellier et al., 2003). The unigenes homologous to HSP90α/β, HSP70, HSP60, and small HSPs (HSP40, HSP27, HSP26, HSP22, and HSP10) were identified in the present study, indicating that heat shock proteins express abundantly and probably play major roles in innate immunity in the foot tissue (Table S5). For the antioxidant enzymes, eight unigenes were homologous to superoxide dismutase (SOD). Three unigenes were similar to catalase (CAT). Six unigenes were homologous to glutathione peroxidase (GPx). Four unigenes were similar to thioredoxin (TRx) and four to TRx-like protein. One unigene was homologous to thioredoxin peroxidase (TPx). One unigene was similar to glutathione reductase (GR). Twenty-five unigenes were homologous to glutathione-S-transferase (GST). These unigenes make up a relatively completely spectrum of antioxidant enzymes, strongly supporting the hypothesis that antioxidant enzymes widely participate in the management of oxidative stress in the P. undulata foot tissue. Lysozymes can cause bacterial cell lysis (Bachali et al., 2002). In this study, five unigenes each were similar with chicken-type, goose-type, and phage-type that were defined by Wang et al. (2013), while two unigenes were homologous to the lysozymes found in other bivalve species (Moreira et al., 2012). The abundant expression of lysozymes may indicate major roles for lysozymes in the P. undulate foot tissue. Mucins (MUC) are large, highly glycosylated viscoelastic macromolecular proteins. In the present study, three unigenes were homologous to the secreted mucins, MUC2 and MUC5AC; four unigenes were similar to the membrane-associated mucins, MUC1, MUC4, MUC16, and MUC22. In addition, six unigenes were homologous to the integumentary mucin C1. To our knowledge, there are few reports concerning mucins in clam species. In P. undulata, the abundant expression of mucins suggests that they probably play an important protective role by forming an immunological barrier between environment and organism as reported in vertebrate (Gendler and Spicer, 1995).
4.3 SSR development and polymorphism validationSSR discovery from high throughput transcriptome data is more effective than the traditional approaches, because the assembly of unigenes is a rich resource for SSR detection and discovery (An and Lee, 2012). In the present study, 7.5% of unigenes possessed SSR loci, which was higher than those of the hard clam Meretrix meretrix (3.1%) and pearl oyster Pinctada maxima (1.6%) and was lower than those of P. textile (8.73%) (Li et al., 2011; Deng et al., 2014; Chen et al., 2016). However, many other SSRs failed in PCR amplification. The location of primers across splice sites, chimeric primers and poor quality sequences might be the main reasons for the failed amplification. Nevertheless, those polymorphic loci identified by this study will be beneficial to the studies of germplasm assessment, quantitative trait loci mapping, and comparative genomics in P. undulata.
5 CONCLUSIONFor the first time, a transcriptome of the foot tissue was obtained using the Illumina Hiseq 2000 sequencing platform, yielding 5.29 Gb raw data in total, and the reads were de novo assembled into 60 201 unigenes with an N50 of 992 bp. BLAST searches against public databases resulted in 22 260 unigenes annotated with significant similarity. Twelve genes that were in response to stress and immune expressed abundantly, suggesting a constant and broad occurrence of immune defense in the foot tissue of P. undulata. Furthermore, 566 unigenes matched to the genes involving in the innate immune reaction system, including exogenous substance discrimination, signal transduction, and immune effectors, indicating that a relatively complete immune mechanism also occurred in the foot tissue. In addition, 16 SSRs developed from the unigenes were polymorphic. The transcriptome analysis could facilitate research about the molecular mechanism of physiological adaptation to tropical environments and the mechanism of innate immunity occurred in foot tissue of P. undulata.
6 DATA AVAILABILITY STATEMENTThe data that supports the findings of this study is submitted to the National Center for Biotechnology Information Short Read Archive, accession numbers SAMN09771584.
Electronic supplementary materialSupplementary material (Supplementary Tables S1–S5 and Figs.S1–S8) is available in the online version of this article at https://doi.org/10.1007/s00343-019-8154-5.
Aguirre J, Ríos-Momberg M, Hewitt D, Hansberg W. 2005. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol., 13(3): 111-118.
DOI:10.1016/j.tim.2005.01.007 |
Akira S. 2003. Toll-like receptor signaling. J. Biol. Chem., 278(40): 38 105-38 108.
DOI:10.1074/jbc.R300028200 |
Altschul S F, Madden T L, Schäffer A, Zhang J H, Zhang Z, Miller W, Lipman D J. 1997. Gapped BLAST and PSI-BLAST:a new generation of protein database search programs. Nucleic Acids Res., 25(17): 3 389-3 402.
DOI:10.1093/nar/25.17.3389 |
An H S, Lee J W. 2012. Development of microsatellite markers for the Korean mussel, Mytilus coruscus (Mytilidae)using next-generation sequencing. Int. J. Mol. Sci., 13(8): 10 583-10 593.
DOI:10.3390/ijms130810583 |
Bachali S, Jager M, Hassanin A, Schoentgen F, Jollès P, Fiala-Medioni A, Deutsch J S. 2002. Phylogenetic analysis of invertebrate lysozymes and the evolution of lysozyme function. J. Mol. Evol., 54(5): 652-664.
DOI:10.1007/s00239-001-0061-6 |
Bahia A C, Kubota M S, Tempone A J, Pinheiro W D, Tadei W P, Secundino N F C, Traub-Csekö Y M, Pimenta P F P. 2010. Anopheles aquasalis infected by Plasmodium vivax displays unique gene expression profiles when compared to other malaria vectors and plasmodia. PLoS One, 5(3): e9795.
DOI:10.1371/journal.pone.0009795 |
Beaz-Hidalgo R, Balboa S, Romalde J L, Figueras M J. 2010. Diversity and pathogenecity of Vibrio species in cultured bivalve molluscs. Environ. Microbiol. Rep., 2(1): 34-43.
DOI:10.1111/j.1758-2229.2010.00135.x |
Bettencourt R, Pinheiro M, Egas C, Gomes P, Afonso M, Shank T, Santos R S. 2010. High-throughput sequencing and analysis of the gill tissue transcriptome from the deep-sea hydrothermal vent mussel Bathymodiolus azoricus. BMC Genomics, 11: 559.
DOI:10.1186/1471-2164-11-559 |
Botstein D, White R L, Skolnick M, Davis R W. 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum.Genet., 32(3): 314-331.
|
Brun N T, Bricelj V M, MacRae T H, Ross N W. 2008. Heat shock protein responses in thermally stressed bay scallops, Argopecten irradians, and sea scallops, Placopecten magellanicus. J. Exp. Mar. Biol. Ecol., 358(2): 151-162.
DOI:10.1016/j.jembe.2008.02.006 |
Carroll M C. 2004. The complement system in regulation of adaptive immunity. Nat. Immunol., 5(10): 981-986.
DOI:10.1038/ni1113 |
Chen X M, Li J K, Xiao S J, Liu X D. 2016. De novo assembly and characterization of foot transcriptome and microsatellite marker development for Paphia textile. Gene, 576(1): 537-543.
DOI:10.1016/j.gene.2015.11.001 |
Clark M S, Thorne M A S, Vieira F A, Cardoso J C R, Power D M, Peck L S. 2010. Insights into shell deposition in the Antarctic bivalve Laternula elliptica:gene discovery in the mantle transcriptome using 454 pyrosequencing. BMC Genomics, 11: 362.
DOI:10.1186/1471-2164-11-362 |
Conesa A, Götz S, García-Gómez J M, Terol J, Talón M, Robles M. 2005. Blast2GO:a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 21(18): 3 674-3 676.
DOI:10.1093/bioinformatics/bti610 |
Cronin S J, Nehme N T, Limmer S, Liegeois S, Pospisilik J A, Schramek D, Leibbrandt A, Simoes R D M, Gruber S, Puc U, Ebersberger I, Zoranovic T, Neely G G, von Haeseler A, Ferrandon D, Penninger J M. 2009. Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science, 325(5938): 340-343.
DOI:10.1126/science.1173164 |
Delanghe J R, Speeckaert R, Speeckaert M M. 2014. Complement C3 and its polymorphism:biological and clinical consequences. Pathology, 46(1): 1-10.
|
Deng Y W, Lei Q N, Tian Q L, Xie S H, Du X D, Li J H, Wang L Q, Xiang Y X. 2014. De novo assembly, gene annotation, and simple sequence repeat marker development using Illumina paired-end transcriptome sequences in the pearl oyster Pinctada maxima. Biosci. Biotechnol. Biochem., 78(10): 1 685-1 692.
DOI:10.1080/09168451.2014.936351 |
Dunkelberger J R, Song W C. 2010. Complement and its role in innate and adaptive immune responses. Cell Res., 20(1): 34-50.
|
Feldmeyer B, Wheat C W, Krezdorn N, Rotter B, Pfenninger M. 2011. Short read Illumina data for the de novo assembly of a non-model snail species transcriptome(Radix balthica, Basommatophora, Pulmonata), and a comparison of assembler performance. BMC Genomics, 12: 317.
DOI:10.1186/1471-2164-12-317 |
Feng B B, Dong L L, Niu D H, Meng S S, Zhang B, Liu D B, Hu S N, Li J L. 2010. Identification of immune genes of the Agamaki clam (Sinonovacula constricta) by sequencing and bioinformatic analysis of ESTs. Mar.Biotechnol., 12(3): 282-291.
DOI:10.1007/s10126-009-9216-z |
Gendler S J, Spicer A P. 1995. Epithelial mucin genes. Annu.Rev. Physiol., 57: 607-634.
DOI:10.1146/annurev.ph.57.030195.003135 |
Gerdol M, Manfrin C, De Moro G, Figueras A, Novoa B, Venier P, Pallavicini A. 2011. The C1q domain containing proteins of the Mediterranean mussel Mytilus galloprovincialis:a widespread and diverse family of immune-related molecules. Dev. Comp. Immunol., 35(6): 635-643.
DOI:10.1016/j.dci.2011.01.018 |
Gourdine J P, Smith-Ravin E J. 2007. Analysis of a cDNA-derived sequence of a novel mannose-binding lectin, codakine, from the tropical clam Codakia orbicularis. Fish Shellfish Immunol., 22(5): 498-509.
DOI:10.1016/j.fsi.2006.06.013 |
Grieshaber M K, Hardewig I, Kreutzer U, Pörlner H O. 1994.Physiological and metabolic responses to hypoxia in invertebrates. In: Reviews of Physiology, Biochemistry and Pharmacology. Springer, Berlin, Heidelberg. p.43-147.
|
Gueguen Y, Cadoret J P, Flament D, Barreau-Roumiguière C, Girardot A L, Garnier J, Hoareau A, Bachère E, Escoubas J M. 2003. Immune gene discovery by expressed sequence tags generated from hemocytes of the bacteria-challenged oyster, Crassostrea gigas. Gene, 303: 139-145.
DOI:10.1016/S0378-1119(02)01149-6 |
Haas B J, Papanicolaou A, Yassour M, Grabherr M, Blood P D, Bowden J, Couger M B, Eccles D, Li B, Lieber M, MacManes M D, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey C N, Henschel R, LeDuc R D, Friedman N, Regev A. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc., 8(8): 1 494-1 512.
DOI:10.1038/nprot.2013.084 |
Huan P, Wang H X, Liu B Z. 2012. Transcriptomic analysis of the clam Meretrix meretrix on different larval stages. Mar.Biotechnol., 14(1): 69-78.
DOI:10.1007/s10126-011-9389-0 |
Iwanaga S, Lee B L. 2005. Recent advances in the innate immunity of invertebrate animals. J. Biochem. Mol. Boil., 38(2): 128-150.
|
Iwasaki A, Medzhitov R. 2004. Toll-like receptor control of the adaptive immune responses. Nat. Immunol., 5(10): 987-995.
DOI:10.1038/ni1112 |
Janeway C A Jr, Medzhitov R. 2002. Innate immune recognition. Annu. Rev. Immunol., 20: 197-216.
DOI:10.1146/annurev.immunol.20.083001.084359 |
Jensen K T, Castro N F, Bachelet G. 1999. Infectivity of Himasthla spp. (Trematoda) in cockle (Cerastoderma edule) spat. J. Mar. Biol. Assoc. UK, 79(2): 265-271.
DOI:10.1017/S0025315498000290 |
Jeong E, Lee J Y. 2011. Intrinsic and extrinsic regulation of innate immune receptors. Yonsei Med. J., 52(3): 379-392.
DOI:10.3349/ymj.2011.52.3.379 |
Jiang WN, Hon Y, Inouye M. 1997. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem., 272(1): 196-202.
DOI:10.1074/jbc.272.1.196 |
Kim J Y, Adhya M, Cho S K, Choi K S, Cho M. 2008. Characterization, tissue expression, and immunohistochemical localization of MCL3, a C-type lectin produced by Perkinsus olseni-infected Manila clams (Ruditapes philippinarum). Fish Shellfish Immunol., 25(5): 598-603.
DOI:10.1016/j.fsi.2008.07.015 |
Lauckner G. 1983. Diseases of Mollusca: Bivalvia. In: Kinne O ed. Diseases of Marine Animals. Biologische Anstalt Helgoland, Hamburg, Germany. p.477-961.
|
Leethochavalit S, Chalermwat K, Upatham E S, Choi K S, Sawangwong P, Kruatrachue M. 2004. Occurrence of Perkinsus sp. in undulated surf clams Paphia undulata from the Gulf of Thailand. Dis. Aquat. Org., 60(2): 165-171.
|
Li H J, Liu W D, Gao X G, Zhu D, Wang J, Li Y F, He C B. 2011. Identification of host-defense genes and development of microsatellite markers from ESTs of hard clam Meretrix meretrix. Mol. Biol. Rep., 38(2): 769-775.
DOI:10.1007/s11033-010-0165-4 |
Loker E S, Adema C M, Zhang S M, Kepler T B. 2004. Invertebrate immune systems- not homogeneous, not simple, not well understood. Immunol. Rev., 198(1): 10-24.
DOI:10.1111/imr.2004.198.issue-1 |
Matsubara H, Ogawa T, Muramoto K. 2006. Structures and functions of C-type lectins in marine invertebrates. Tohoku J. Agric. Res., 57(1-2): 71-86.
|
Moreira R, Balseiro P, Planas J V, Fuste B, Beltran S, Novoa B, Figueras A. 2012. Transcriptomics of in vitro immune-stimulated hemocytes from the Manila clam Ruditapes philippinarum using high-throughput sequencing. PLoS One, 7(4): e35009.
DOI:10.1371/journal.pone.0035009 |
Mouritsen K N, Poulin R. 2003. The risk of being at the top:foot-cropping in the New Zealand cockle Austrovenus stutchburyi. J. Mar. Biol. Assoc. UK, 83(3): 497-498.
DOI:10.1017/S0025315403007409h |
Niu D H, Jin K, Wang L, Feng B B, Li J L. 2013b. Molecular characterization and expression analysis of four cathepsin L genes in the razor clam, Sinonovacula constricta. Fish Shellfish Immunol., 35(2): 581-588.
DOI:10.1016/j.fsi.2013.06.001 |
Niu D H, Wang L, Sun F Y, Liu Z J, Li J L. 2013a. Development of molecular resources for an intertidal clam, Sinonovacula constricta, using 454 transcriptome sequencing. PLoS One, 8(7): e67456.
DOI:10.1371/journal.pone.0067456 |
O'Connell-Milne S, Poulin R, Savage C, Rayment W. 2016. Reduced growth, body condition and foot length of the bivalve Austrovenus stutchburyi in response to parasite infection. J. Exp. Mar. Biol. Ecol., 474: 23-28.
DOI:10.1016/j.jembe.2015.09.012 |
Pallavicini A, del Mar Costa M, Gestal C, Dreos R, Figueras A, Venier P, Novoa B. 2008. High sequence variability of myticin transcripts in hemocytes of immune-stimulated mussels suggests ancient host-pathogen interactions. Dev.Comp. Immunol., 32(3): 213-226.
DOI:10.1016/j.dci.2007.05.008 |
Parcellier A, Gurbuxani S, Schmitt E, Solary E, Garrido C. 2003. Heat shock proteins, cellular chaperones that modulate mitochondrial cell death pathways. Biochem.Biophys. Res. Commun., 304(3): 505-512.
DOI:10.1016/S0006-291X(03)00623-5 |
Paul-Pont I, de Montaudouin X, Gonzalez P, Soudant P, Baudrimont M. 2010. How life history contributes to stress response in the Manila clam Ruditapes philippinarum. Environ. Sci. Pollut. Res., 17(4): 987-998.
DOI:10.1007/s11356-009-0283-5 |
Peng M X, Niu D H, Wang F, Chen Z Y, Li J L. 2016. Complement C3 gene:Expression characterization and innate immune response in razor clam Sinonovacula constricta. Fish Shellfish Immunol., 55: 223-232.
DOI:10.1016/j.fsi.2016.05.024 |
Philipp E, Pörtner HO, Abele D. 2005. Mitochondrial ageing of a polar and a temperate mud clam. Mech. Ageing Dev., 126(5): 610-619.
DOI:10.1016/j.mad.2005.02.002 |
Ruddell R G, Hoang-Le D, Barwood J M, Rutherford P S, Piva T J, Watters D J, Santambrogio P, Arosio P, Ramm G A. 2009. Ferritin functions as a proinflammatory cytokine via iron-independent protein kinase C zeta/nuclear factor kappaB-regulated signaling in rat hepatic stellate cells. Hepatology, 49(3): 887-900.
DOI:10.1002/hep.22716 |
Sambrook J, Fritsch E F, Maniatis J. 1989. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor Laboratory Press, New York. 1625p.
|
Shi M J, Lin Y, Xu G R, Xie L P, Hu X L, Bao Z M, Zhang R Q. 2013. Characterization of the Zhikong scallop(Chlamys farreri) mantle transcriptome and identification of biomineralization-related genes. Mar. Biotechnol., 15(6): 706-715.
DOI:10.1007/s10126-013-9517-0 |
Song L S, Wang L L, Qiu L M, Zhang H A. 2011. Bivalve immunity. In: Söderhäll K ed. Invertebrate Immunity.Springer, Boston, MA. p.44-65.
|
Song X Y, Zhang H, Zhao J M, Wang L L, Qiu L M, Mu C K, Liu X L, Qiu L H, Song L S. 2010. An immune responsive multidomain galectin from bay scallop Argopectens irradians. Fish Shellfish Immunol., 28(2): 326-332.
DOI:10.1016/j.fsi.2009.11.016 |
Takeda K, Akira S. 2005. Toll-like receptors in innate immunity. Int. Immunol., 17(1): 1-14.
|
Tatusov R L, Galperin M Y, Natale D A, Koonin E V. 2000. The COG database:a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res., 28(1): 33-36.
DOI:10.1093/nar/28.1.33 |
Thieringer H A, Jones P G, Inouye M. 1998. Cold shock and adaptation. Bioessays, 20(1): 49-57.
DOI:10.1002/(SICI)1521-1878(199801)20:1<>1.0.CO;2-8 |
Thomas F, Renaud F, de Meeûs T, Poulin R. 1998. Manipulation of host behaviour by parasites:ecosystem engineering in the intertidal zone? Proc. Roy. Soc. B:Biol. Sci., 265(1401): 1 091-1 096.
DOI:10.1098/rspb.1998.0403 |
Underhill D M, Ozinsky A. 2002. Toll-like receptors:key mediators of microbe detection. Curr. Opin. Immunol., 14(1): 103-110.
|
van de Wetering J K, van Golde L M G, Batenburg J J. 2004. Collectins:players of the innate immune system. FEBS J., 271(7): 1 229-1 249.
|
Venier P, Varotto L, Rosani U, Millino C, Celegato B, Bernante F, Lanfranchi G, Novoa B, Roch P, Figueras A, Pallavicini A. 2011. Insights into the innate immunity of the Mediterranean mussel Mytilus galloprovincialis. BMC Genomics, 12: 69.
DOI:10.1186/1471-2164-12-69 |
Wang A M, Wang Y, Gu Z F, Li S F, Shi Y H, Guo X M. 2011. Development of expressed Sequence tags from the pearl oyster, Pinctada martensii Dunker. Mar. Biotechnol., 13(2): 275-283.
DOI:10.1007/s10126-010-9296-9 |
Wang L L, Qiu L M, Zhou Z, Song L S. 2013. Research progress on the mollusc immunity in China. Dev. Comp.Immunol., 39(1-2): 2-10.
DOI:10.1016/j.dci.2012.06.014 |
Wang L L, Song L S, Zhao J M, Qiu L M, Zhang H, Xu W, Li H L, Li C H, Wu L T, Guo X M. 2009. Expressed sequence tags from the Zhikong scallop (Chlamys farreri):discovery and annotation of hostdefense genes. Fish Shellfish Immunol., 26(5): 744-750.
DOI:10.1016/j.fsi.2009.03.002 |
Weber M H W, Fricke I, Doll N, Marahiel M A. 2002. CSDBase:an interactive database for cold shock domaincontaining proteins and the bacterial cold shock response. Nucleic Acids Res., 30(1): 375-378.
DOI:10.1093/nar/30.1.375 |
Wu X W, Tan J, Cai M Y, Liu X D. 2014. Molecular cloning, characterization, and expression analysis of a heat shock protein (HSP) 70 gene from Paphia undulata. Gene, 543(2): 275-285.
DOI:10.1016/j.gene.2013.11.103 |
Yang C Y, Wang L L, Siva V S, Shi X W, Jiang Q F, Wang J J, Zhang H, Song L S. 2012. A novel cold-regulated cold shock domain containing protein from scallop Chlamys farreri with nucleic acid-binding activity. PLoS One, 7(2): e32012.
DOI:10.1371/journal.pone.0032012 |
Yeh F C, Yang R, Boyle T J, Ye Z, Xiyan J M. 2000. PopGene32, Microsoft Windows-based freeware for population genetic analysis, version 1.32. Molecular Biology and Biotechnology Centre, University of Alberta, Edmonton, Alberta, Canada.
|
Yeh H Y, Klesius P H. 2010. Characterization and tissue expression of channel catfish (Ictalurus punctatus Rafinesque, 1818) ubiquitin carboxyl-terminal hydrolase L5 (UCHL5) cDNA. Mol. Biol. Rep., 37(3): 1 229-1 234.
DOI:10.1007/s11033-009-9493-7 |
Zhang H, Song L S, Li C H, Zhao J M, Wang H, Qiu L M, Ni D J, Zhang Y. 2008. A novel C1q-domain-containing protein from Zhikong scallop Chlamys farreri with lipopolysaccharide binding activity. Fish Shellfish Immunol., 25(3): 281-289.
DOI:10.1016/j.fsi.2008.06.003 |
 2019, Vol. 37
2019, Vol. 37




