Institute of Oceanology, Chinese Academy of Sciences
Article Information
- GUO Li, YANG Guanpin
- Nannochloropsis artificial chromosomes (NannoACs) loom on the horizon
- Journal of Oceanology and Limnology, 41(6): 2336-2347
- http://dx.doi.org/10.1007/s00343-022-2302-z
Article History
- Received Aug. 30, 2022
- accepted in principle Oct. 13, 2022
- accepted for publication Oct. 24, 2022
2 Institute of Evolution and Marine Biodiversity, Ocean University of China, Qingdao 266003, China;
3 Key Laboratory of Marine Genetics and Breeding of Ministry of Education, Ocean University of China, Qingdao 266003, China
The genus Nannochloropsis comprises seven species that inhabit seawater, freshwater and brackish (Hibberd, 1981; Fawley and Fawley, 2007). The genus has been split into Nannochloropsis and Microchloropsis according to the differences of the concatenated 18S ribosomal RNA and rbcL genes (Fawley et al., 2015). Either in Nannochloropsis or Microchloropsis, all the species are assigned to class Eustigmatophyceae, phylum Ochrophyta, superphylum Heterokonta (or Stramenopiles), which are extremely small, nonmotile and spherical in shape (Bailey and Freshwater, 1997; Daugbjerg and Andersen, 1997; Andersen et al., 1998; Kandilian et al., 2013). These species are monoploidy and asexual (Galloway, 1990; Pan et al., 2011). Nannochloropsis species are able to accumulate high concentrations of pigments including astaxanthin, zeaxanthin, and canthaxanthin (Lubián et al., 2000) and have chlorophyll a but completely lack chlorophylls b and c (Manning and Strain, 1943). Nannochloropsis species are mainly used as feed for fish larvae and rotifers and as food additives for human consumption. They also promise to be industrially utilized because they accumulate a large quantity of polyunsaturated fatty acids (Boussiba et al., 1987; Sukenik et al., 1989).
Species in genus Nannochloropsis have been evolving into the models for both industrial applications and biological researches (Gee and Niyogi, 2017). The genomes of two Nannochloropsis species, N. gaditana (Radakovits et al., 2012; Carpinelli et al., 2014) and N. oceanica (Pan et al., 2011; Vieler et al., 2012; Liang et al., 2013; Guo et al., 2019; Gong et al., 2020), have been sequenced, which vary between 28.5 and 29.3 Mb in size. A phylogenetic analysis of Nannochloropsis species genomes revealed a significant dose expansion among lipid biosynthesis genes (Wang et al., 2014). The molecular tools including genetic transformation (Li et al., 2014; Chen and Hu, 2019; Abidin et al., 2020), targeted gene replacement by homologous recombination (Kilian et al., 2011), CRISPR/Cas9-based genome edition (Wang et al., 2016; Poliner et al., 2018c; Naduthodi et al., 2019; Zhang et al., 2019), gene stacking (Verruto et al., 2018; Poliner et al., 2020), transient expression of genes in episomes (Poliner et al., 2018b; Kurita et al., 2020; Wang et al., 2021), and sets of reporters and selection markers (Poliner et al., 2018b; Naduthodi et al., 2021b) have been rapidly developing in Nannochloropsis. Unfortunately, the genes, either alone or being stacked, are not able to express sustainably if not integrated into genome and cause the position effect if integrated into genome. In addition, the genetic modification of microalgae, for example, the enhancement of productivity and lipid production (Muñoz et al., 2021; Naduthodi et al., 2021a; Zulaiha, 2021; Chen et al., 2022), cannot detour genetically engineering complex traits, metabolic pathways among others and combining genes locating separately in genome. Artificial chromosomes should aid to synthesize (integrate) separate genes and allow us to modify complex traits and metabolic pathways. The expressions of concatenate genes and the genes in large chromosomal fragments appreciate developing Nannochloropsis artificial chromosomes (NannoACs).
2 CURRENT STATUS OF THE GENE ENGINEERING OF NANNOCHLOROPSIS 2.1 Expression of single genesThe expression of a single gene may modify the trait of Nannochloropsis species. For example, the NobZIP77 of N. oceanica represses the transcription of a type-2 diacylgycerol acyltransferase gene under nitrogen-repletion condition. However, it relieves such repression under nitrogen-depletion condition. In addition, it senses blue light, and changes the growth rate and fatty acid productivity of the alga once being inactivated (Zhang et al., 2022). The expression of a transgene may be silenced due to its integration into the host genome (position effect). To detour the genomic integration, a "safe harboring" strategy has been established for Nannochloropsis. The best integration position in genome is determined first according to the expression intensiveness of a reporter gene, and then the transgene is harbored (knocked in) this position with the aid of Cas9-mediated edition. Fatty acid desaturase (FAD12) gene encoding Δ12-fatty acid desaturase at this preferred locus significantly increased its expression and changed the fatty acid composition (Ryu et al., 2021). Recently, Südfeld et al. (2022) fortunately discovered a strong gene expression site within Nannochloropsis nucleolus where the cellular rRNA synthesis machinery is recruited for transcription. The exploited by the expression system include the highly efficient transcriptional activity of RNA polymerase I and an internal ribosome entry site for translation. Successful genetic/metabolic engineering of Nannochloropsis species has improved their phenotypes including, for example, violaxanthin yield (Park et al., 2021) and growth rate and lipid content (Ma et al., 2016). Unfortunately, the evaluation of influence-less position in genome is time consuming and labor intensive. It is hard to popularize such strategy in our routine trying to express foreign genes.
2.2 Stacking genes for metabolism pathwaysThe common situation we encounter often is that a metabolite synthesis pathway, a trait among others are run by a set of genes. Engineering these pathways and traits often requires the co-expression of multiple transgenes. To meet this requirement, a strategy called "gene stacking" has been established in N. oceanica CCMP1779. A set of gateway entry and destination vectors are developed first, and then the entry cassette is stacked onto the destination vector via the phiC31-att integration system (Reece-Hoyes and Walhout, 2018; Poliner et al., 2020). As an early trial, the enzymes involved in long-chain polyunsaturated fatty acid (LC-PUFA) biosynthesis in N. oceanica CCMP1779 are identified and multigene expression vectors are generated to increase LC-PUFA content in vivo. The cDNAs encoding four fatty acid desaturases are isolated, which are then stacked together. The over expressions of these genes in different combinations lead to an increase in eicosapentaenoic acid (EPA) content (Poliner et al., 2018c). "Gene stacking" is not a new term and a new idea in plant breeding, which has been using in the context of genetically modified crops for many years (Halpin, 2005). Plant breeders always stack genes by crossing parents each carrying a desirable trait and then identifying offspring inheriting both of these desired traits.
Gene stacking appreciates the improvement of the gene carrying capability of vectors. The carrying capability of a plasmid vector cannot be expanded unlimitedly. One of the solutions is to adopt the bacterial artificial chromosome (BAC) which may carry a large number of genes in both fusion protein and concatenate gene cassettes and their expression controlling elements. A BAC is able to clone DNA sequences varying from 100 to about 300 kb (Shizuya and Kouros-Mehr, 2001; Zhang et al., 2021). Except for the limitation in the numbers of controlling elements, reporter genes and selection markers, the influence of integration position in the genome is also a challenge (Ryu et al., 2021). Plasmids are used in bacteria to build up gene constructs. Upon transformed into microalgae, they must integrate into the chromosomes, thus are called integrating vectors. Although they can deliver genetic materials to a target cell and enable long-term expression of transgenes, their integration derives a potential risk of causing insertional mutagenesis. In contrast, episomal vectors behave as separate extrachromosomal elements, thus may avoid these undesired side effects (Ehrhardt et al., 2008). The insertional mutation may disrupt the genes anywhere in host genome while the expression of genes on episomal vector does not interrupt any gene due to their independent extrachromosomal existence. Without selection pressure of antibiotic markers, it will disappear through biological dilution.
2.3 Non-integrative vectorEpisomal vectors may aid to overcome the disadvantages of integrative ones. Very interestingly, foreign CEN and ARS have been proved to function in N. oceanica, and a one-vector CRISPR system for non-integrative gene disruption in N. oceanica has been developed (Poliner et al., 2018c). A hygromycin B resistance cassette, a Cas9-3×glycine-serine linker-reporter fusion protein gene promoted by a bidirectional promoter and terminated by LDSP terminator and the sgRNA promoted by the same bidirectional promoter and terminated by a CS terminator are expressed simultaneously. The fusion protein is assigned into nucleus by placing SV40 nuclear localization signals at its N' and C' termini. The sgRNA is trimmed automatically after transcription by self-cleaving 3' HDV and 5' HH ribozymes. The Saccharomyces cerevisiae CEN/ARS6 region is recombined into the episome to create an episomal CRISPR system which will lose when selection pressure is removed. This design assures the edited cells to be marker-free. The circular plasmids containing CEN/ARS are not inserted into the chromosome and can be removed after editing (Kurita et al., 2022). The nuclear episomal vector for gene cloning in diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana has also been developed (Karas et al., 2015). The episome is not stable; it will be diluted when the antibiotics is removed away. The episome is the choice for gene edition; however, it is not appropriate for transgenes.
2.4 The prospect of Nannochloropsis YAC-like vectors looks bleakA method similar to the construction of yeast artificial chromosome (YAC) could be used to develop microalgal artificial chromosomes (Fabris et al., 2020). Actually, only one-step ahead to evolve episome towards the YAC-like vectors in Nannochloropsis. Four essential chromosomal elements, a yeast centromere CEN4, an autonomously replicating sequence ARS1 and two Tetrahymena telomeric sequences TELs, function in yeast, making YAC replicate and vertically transfer from parents to daughter cells as a real yeast chromosome does. YAC can be modified thorough homologous recombination to carry additional selection markers and genes or delete the unwanted regions or recombine with other YAC (Burke et al., 1987; Arnak et al., 2012).
Embedding animal cells and plant protoplasts in agarose plugs and then extracting DNA within the agarose plugs minimize the risk of DNA breaking. The extracted DNA can be stored for long periods, partially fragmented enzymatically (Nair et al., 1999), and separated through pulsed-field gel electrophoresis (PFGE) (Hicks et al., 2018). Once being inserted into a YAC, the extra-long DNA fragments can be cloned and amplified in yeast. The spectrum of YAC applications is very wide, from constructing DNA libraries of the large genomes to identifying genes within a large chromosomal region. YAC can also be retrofitted with the desired genes and appropriate selective markers and transferred into animal cells, with the functions of genes within a large DNA region deciphered there. In addition, the selection markers of YAC are the normal genes of an auxotrophic mutant.
To build up a construct containing CEN, ARS, and TEL cassettes may detour plasmid and bacteria by polishing (or blunting) and ligating DNA fragments and PCR products of CEN, ARS, and TEL elements, or via synthetic sequence-guided isothermal assembly (Torella et al., 2014; Kadkhodaei et al., 2016). As found in practice, CEN/ARS of yeast does not allow the episomal vectors of Nannochloropsis to have enough stability. Although not proved, YAC may not be transplanted into Nannochloropsis. We must consider the stability of the yeast CEN/ARS6 region in episomal vectors of Nannochloropsis, since the vector may lose when selection pressure is removed (Kurita et al., 2022).
3 ANIMAL AND PLANT ARTIFICIAL CHROMOSOME EXAMPLES OF NANNOACSIn contrast, mammalian artificial chromosomes, for example, human artificial chromosomes (HACs) and mouse artificial chromosomes (MACs), can carry multiple genes or megabase-sized genomic loci and their associating regulatory elements, facilitate creating more useful and complex transgenic models for human disease studies and drug development (Moriwaki et al., 2020). HACs and MACs have set fine examples for the expecting NannoACs. Two strategies, "top down" and "bottom up", have been used to develop HACs and MACs.
3.1 HAC and MAC constructions with "top down" strategyThe P1 bacteriophage cyclization recombination recombinase (Cre) mediates the recombination between paired lox sites. Depending on the promoters and other regulatory controls, Cre in bacteria, fungi, plants, and animals can be manipulated to synthesize Cre recombinase under certain conditions. Cre can also be expressed transiently by transfecting eukaryotic cells with bacterial plasmid containing Cre and its controlling elements. The flanked lox sites integrated in eukaryotic chromosomes may cause deletions, inversions and translocations when Cre recombinase is available, which is called Cre-lox recombination system. Similarly, FLP-frt system is becoming more frequently used in mouse-based researches, in which flippase (FLP) recombinase derived from the yeast S. cerevisiae recognizes and recombines paired FLP recombinase target (frt) motifs that flank a genomic region of interest (Araki et al., 1997; Schweizer, 2003).
The top-down construction of a HAC needs to reduce the size of a natural chromosome by targeted truncation. In the chicken DT40 cells allowing a high rate of homologous recombination, the transferred human chromosome 22 was truncated at LIF locus and ligated with human telomeric repeats (TTAGGG)n (Kuroiwa et al., 1998). Homologous recombination based chromosomal truncation in eukaryotic cells provides the opportunity of constructing HACs with a "top-down" strategy and expressing genes in an extremely large genetic locus. The HAC is transferred into mouse embryo stem cells that are then injected into host embryo, leading to generate chimeric and germline transmissible mouse (Tomizuka et al., 1997). With the aids of lox and Cre recombinase, defined regions of human chromosomes > one megabase in length can be cloned into a stable HAC in homologous recombination-proficient chicken DT40 cells (Kuroiwa et al., 2000). With the "top-down" strategy and the homologous recombination systems, a MAC with essential centromere and telomeres was constructed, which contains human Ig heavy chain locus (IGH) derived from hChr.14 and Ig kappa light chain locus (IGK) derived from hChr.2. It is maintained in transchromosomic mice which produces human antibody in the mouse Ig-knockout background (Satofuka et al., 2022).
With the "top-down" strategy, MACs from native mouse chromosomes 10 and 11 (10MAC and 11 MAC) have been constructed, which carry diverse markers like puromycin and neomycin resistances, enhanced green fluorescent protein (GFP) among others and Cre-lox DNA loading system (Takiguchi et al., 2014; Abe et al., 2021). HAC derived from native human chromosomes, independently and stably maintained without disrupting host chromosomes, have been also constructed, which carry recombination sites for loading genes, and are capable to transfer a large quantity of megabase sized genes (Oshimura et al., 2015).
3.2 HAC and MAC construction with "bottom up" strategyIn contrast to "top-down construction", the "bottom-up construction" strategy involves the de novo buildup of HACs and MACs by introducing DNA elements necessary for maintaining chromosome function into cells. A HAC is a mini-chromosome behaving as a stable chromosome in host nucleus using its own self-replicating and segregating systems. Using the "bottom-up" strategy, a HAC is constructed by transfecting a cultured human cell line (for example, HT1080) with cloned or synthetic centromeric alphoid DNA precursors with CENP-B boxes, where the precursors multimerize up to megabase size (Ikeno and Suzuki, 2011). Alphoid-BAC containing 50-kb alphoid DNA and lox-BAC containing the CAG/lox71/neo cassette with insulator elements, 5'HS5 and 3'HS1 at left and right ends are co-transfected into HT1080 cells, from them the cells with multimerized HAC are subsequently selected out according to neomycin resistance and through fluorescence in-situ hybridization (FISH) analysis. The HAC constructed can be transferred into mouse cells with microcell-mediated chromosome transfer method or transchromosomic technique. Interested genes or DNA fragments in plasmid may be inserted into HAC via Cre-lox system.
A tetracycline response element (TRE) is the 7 repeats of a 19 nucleotides tetracycline operator (tetO) sequence, and is recognized by the tetracycline repressor (TetR). A tetracycline-controlled transactivator (tTA) is created by fusing TetR with the C-terminal domain of virion protein 16 (VP16), an essential transcriptional activation domain from herpes simplex virus (HSV). Tetracycline or its analogs like doxycycline switches off the expression of the genes downstream TRE by binding to tTA (Gossen and Bujard, 1992). Later, the TetR is mutated into a reverse tetracycline repressor rTetR, which induces rather than impress the expression of the genes downstream TRE when tetracycline presents. The new reverse tetracycline-controlled transactivator (rtTA) is created by fusing rTetR with VP16. This bacterial system functions in mammalian cells, making the expression inducible and controllable. Alphoid DNA and its variant with CENP-B box replaced with the tetO motif (Ponomartsev et al., 2020) are circulated and amplified in a way of rolling PCR. The PCR product is inserted in between hooks of a plasmid and transferred into yeast where inserts are assembled by transformation-associated recombination into approximately a 50-kb synthetic alphoid array as a circular molecule. Recombination further multimerizes the array into 1.1 Mb in size, forming the alphoidtetO-HAC, which is proved to be a versatile tool for studying human chromosome transactions and structure as well as for genome and cancer studies (Kouprina et al., 2018). The alphoidtetO-HAC has also been modified to be applicable for assembling (iterate) genomic DNA fragments or genes with regulated kinetochore using three combinations of integrases (recombinase) and mediate motifs, Cre-lox, phiC31-att, and phiBT1-att, making cells capable of carrying multiple transgenes and megabase-sized genomic loci, thus allowing us to investigate complex biomedical pathways and gene regulation, and engineer synthetic chromosomes with a predetermined set of genes (Lee et al., 2018).
In essence, the HACs and MACs constructed with "bottom-up" strategy are the modified centromeres, into which genes can be integrated via the mediate motif and recombinase systems. They are circular DNA thus their sustainable inheritance does not need telomeres. These artificial chromosomes work; however, their replication effeciency is not determined. It is possible that such chromosome may not perform as good as the natural ones. In addition, the transformation efficiency may hinder effectively transferring large DNA constructs into Nannochloropsis cells. To these considerations, as the preliminary trials, it is better to construct NannoACs by truncating natural Nannochloropsis chromosomes, i.e., with the top-down strategy.
3.3 Plant minichromosmes (artificial chromosomes)The artificial chromosomes have also been constructed in plants. Although widely debated publicly, yet genetic engineering with just a few genes, most frequently the herbicide resistance gene for efficient weed control and the Bacillus thuringiensis (Bt) toxin gene for insect resistance, have changed modern crop cultivation. Technical advancements provide more potentials for genetically engineering crops, which include plant artificial chromosome technology. It allows us to manage a large number of genes or extremely long DNA fragments and complex traits. Plant artificial chromosomes (PACs) or plant minichromosomes are usually small in size, which contain only essential chromosome components such as the centromere, telomeres and replication origins in order to maintain the normal chromosome function. Similar to animal artificial chromosomes, PACs can also be constructed with "bottom up" (de-novo assembling) and "top down (natural chromosome truncating) strategies. Comparatively, telomere mediated chromosomal truncation (TMCT) has meet more successes (Yu et al., 2016). TMCT depends on the transformation of a telomere-containing sequence into the genome where it replaces one of the old once integrated into the chromosome (Yu et al., 2006). Between T-DNA borders, bialophos resistance gene, lox and frt, hygromycin B-resistance gene, GFP, red fluorescent protein gene, recombinase gene and telomere units isolated from Arabidopsis thaliana were arrayed in concatenation, and introduced into cells where it merged with a truncated chromosome with the centromere and the telomere at the distant end. Since the establishment of TMCT in maize (Yu et al., 2006), PACs have been constructed in many plant species including maize, rice, Arabidopsis, barley among others.
To manipulate genes or DNA fragments, PACs must contain different recombination systems, such as the Cre-lox (Abremski et al., 1983), FLP-frt (Golic and Lindquist, 1989), and the phiC31-att (Thorpe and Smith, 1998) available currently. With the aid of these recombination systems, the selection marker genes and interested genes can be exchanged between a PAC and the donor, and the desirable genes can be stacked on a PAC (Yu et al., 2016). Instead of artificial chromosome construction, a simple method is to insert a single-copy T-DNA from pDs-Lox into the centromere of an Arabidopsis chromosome. When Ac TPase gene is transformed into and transiently expressed in the plant genome, a cassette flanked by the inverted terminal region of maize Ds is transposed into right or left distant region of the chromosome. Mediated by Cre recombinase, the recombination between lox sites generated PACs into which a gene may be cloned at the lox site (Murata et al., 2013).
4 CONSTRUCTION OF NANNOACS 4.1 Nannochloropsis species possess the prerequisites for NannoAC constructionOf the widely used three recombination systems, Cre-lox (Abremski et al., 1983), FLP-frt (Golic and Lindquist, 1989), and the phiC31-att (Thorpe and Smith, 1998), phiC31-att and Cre-lox systems have been shown to work in Nannochloropsis (Verruto et al., 2018; Poliner et al., 2020). Robust technical tools in microalgae including Nannochloropsis species are essential to manipulate both intrinsic (endogenous) and extrinsic (foreign) genes in their genomes for basic and applied studies. While Cas9-mediated gene editing has been developed in Nannochloropsis species (Wang et al., 2016; Naduthodi et al., 2019, 2021b; Kurita et al., 2022), the molecular tools of knocking in or knocking out "traits" controlled by a group of genes within a large or among a group of DNA fragments are appreciated also. A concatenate construct, pSliceN'Excise, which contains Cas9, nitrite inducible Cre and floxed (flanked by lox sites) blasticidin resistance (BSD) and GFP expression cassettes, has been generated. After integration, the induced Cre eliminates the marker and the reporter, leaving a marker- and reporter-free genome constantly expressing Cas9 and expressing Cre upon induction. In this genome, the gRNA aids to knock out acyl-CoA oxidase gene (Aco1) by inserting the marker and reporter expression cassettes. Co-electroporating the specific gRNA with floxed marker/reporter cassette with additional sequence both sides cause insertion of the marker/reporter during DNA repairing at the Cas9 target site. After Cre induction, the marker/reporter cassette is removed and the additional sequence is stably integrated into and inactivated the ZnCys. By employing Cas9 and Cre, stepwise iterative knocking out and deleting marker/reporter accumulatively modify the multiple genes of a trait (Verruto et al., 2018).
Today, groups of selection markers, reporters, and their expression controlling elements have been identified and used (Poliner et al., 2018a, b, 2020). With these foundations, we may try to construct NannoACs with both "bottom up" and "top down" strategies. As shown in episomal vector of Nannochloropsis, yeast CEN/ARS works but may not assure the stable inheritance of the plasmid. Such scenario may not change when the telomeres are combined. The genomes of Nannochloropsis have been repeatedly sequenced and assembled (Guo et al., 2019; Gong et al., 2020); unfortunately, the telomere to telomere assembly is not available yet. The construction of NannoACs appreciate the sequence of centromeres. The ultralong reads degenerated by Nanopore sequencing make it possible to obtain the sequence of telomeres of Nannochloropsis and then construct NannoACs with bottom up strategy. The time has come to examine the complete sequence of Nannochloropsis centromeres (Miga et al., 2020; Suzuki and Morishita, 2021). With the background information either available today or conveniently obtainable with the existing techniques, we may construct the NannoACs with both bottom-up strategy and top-down strategy that are feasible in both mammals and plants.
Adding an extra chromosome into the nuclei of diploidy animals and plants may not interfere the functioning of nuclei. However, when we construct NannoACs with top-down strategy, the truncation of a chromosome of the monoploidy Nannochloropsis may be lethal. We have found that both mammals and plants tolerating chromosomal truncation are diploids, which allow one of the two homologous chromosomes to be truncated.
The genome of Nannochloropsis is monoploidy (Pan et al., 2011). Although some parts of the genome of Nannochloropsis may not absolutely necessary always (Wang et al., 2021), and plant genomes may allow deletion of some regions (Tan et al., 2020), it is imaginable that truncating any chromosome to some extent will be lethal to the cells. Colchicine induces chromosome doubling of plants (Manzoor et al., 2019). It also induces chromosomal doubling of microalgae (Nezhad and Mansouri, 2019; Szpyrka et al., 2020; Le-Feuvre et al., 2021; Mansouri et al., 2021). The diploid of doubled genomes has been traced for a certain amount of generations; however, whether the diploidy state can be maintained permanently is not sure. In cyanobacteria, the nuclear ploidy tightly associated with the environment (Zerulla et al., 2016; Riaz et al., 2021). It is reasonable to believe that the doubled genome is not permanently stable. As a preliminary trial, we are doubling the chromosomes of N. oceanica and trying to maintain its stability by Zeocin mutation that causes double strand breaks of DNA and introduces insertion and deletion during cellular DSB repairing (Yang et al., 2019). At a locus of doubled thus completely homologous chromosomes, a normal and a lethal allele may locate, surviving both chromosomes through functional complementation. Such scenario should aid to maintain the diploidy genome during cultivation.
Homologous recombination may destroy the maintenance of diploidy genome; however, homologous recombination may happen at an extremely low frequency in somatic cells; it takes place only when the alleles align together as are observed in meiosis. To this consideration, our strategy of diploid maintenance is the mutated nutrition alleles. As an alternative way of diploid maintenance, we may introduce different antibiotic markers into the doubled chromosomes, one each, and maintain the diploid of Nannochloropsis with two antibiotics. Actually, Nannochloropsis species have been verified to perform double antibiotic resistance simultaneously (Poliner et al., 2018b).
4.2 NannoACs loom on the horizonNannochloropsis species are monoploidy (Pan et al., 2011). Some regions of their genomes including telomeres can be deleted (Wang et al., 2021); however, we prefer to believe that all chromosomal regions are necessary for their survival in all possible environments. Therefore, we may double their chromosomes with colchicine. It has been demonstrated that the doubled genome may maintain for a period (Nezhad and Mansouri, 2019; Szpyrka et al., 2020; Le-Feuvre et al., 2021; Mansouri et al., 2021); however, phosphate depletion will certainly reduce the ploidy of cyanobacteria (Zerulla et al., 2016; Riaz et al., 2021). We are endeavoring to mutate the genes essential for Nannochloropsis survival with Zeocin, a double strand breaker of DNA (Chankova et al., 2007; Yang et al., 2019), aiming to maintain the diploidy after chromosomal doubling. The mutated allele will stabilize the doubled chromosomes, allowing us to truncate one of the homologous chromosome pair.
To survive all possible environments, monoploidy Nannochloropsis genome should be doubled through colchicine treatment. The essential genes in doubled chromosomes may be mutated with zeocin so that the functionally complementary alleles can maintain the diploidy karyotype. If three mediate motif-recombinase systems work in Nannochloropsis, constructing a NannoAC with three mediate motifs, one works alone and two work in combination, will be feasible. Simultaneous transformation of the episome carrying editing parts and the telomere constructs consisting of mediate motifs and telomere repeats will generate a NannoAC. With the aids of transiently expressed corresponding recombinases, we may integrate genes, concatenate genes and DNA fragments into NannoACs (see the main text for the details).
Of three combinations of mediate motif-recombinase (Abremski et al., 1983; Golic and Lindquist, 1989; Thorpe and Smith, 1998), Cre-lox and phiC31-att systems have been found to function well in Nannochloropsis (Verruto et al., 2018; Poliner et al., 2020). Once all of the three systems work in Nannochloropsis, it is feasible to construct a NannoAC with three mediate motifs, one works alone and two work in combination. To construct NannoAC with top-down strategy, Nannochloropsis cells with doubled genome are simultaneously transformed with the episome carrying sgRNA targeting selected chromosomal regions and the artificial telomere constructs consisting of mediate motifs and Nannochloropsis telomere repeats (for example, Wang et al., 2021). The genomes of Nannochloropsis species have been sequenced repeatedly (Guo et al., 2019; Gong et al., 2020). With the telomere to telomere (T2T) sequencing technology (Miga et al., 2020), the precise sequences of Nannochloropsis centromeres and telomeres will allow us to construct NannoACs with both "top down" and "bottom up" strategies. The applications of expected NannoACs are diverse. Either one mediate motif or two in combination can mediate the integration of concatenate genes (reporters, selection markers, the interested genes and their expression controlling elements) with the aid of co-transferred recombinase expression cassettes or even recombinases themselves. Reporters and selection markers should aid to screen out the transformants. However, multiplex PCR (Elnifro et al., 2000) and pooling design (Bruno et al., 1995) may accelerate the screening of transformants especially when the concatenate genes or DNA fragment are extremely long. Operating long concatenate genes and large DNA fragments and avoiding position effect are the advantages of artificial chromosomes over integration-based transgene vectors while the stability of artificial chromosomes is the advantage over the episome. To both artificial chromosomes and integration-based vectors, bacterial plasmids and artificial chromosomes provide conveniences for handling genes and DNA fragments. Extralong DNA can be isolated from any cell wall free cells including animal cells and plant protoplasts (Nair et al., 1999) and separated through pulse field gel electrophoresis (Sharma-Kuinkel et al., 2016). Karyotyping microalgae may be difficult (Muravenko et al., 2001; Sánchez-Gárate et al., 2020). Fortunately, many pseudochromosomes (the chromosomal scale assemblies) of Nannochloropsis are < one megabase (Guo et al., 2019; Gong et al., 2020) and the isolation of microalgal protoplasts has been progressing (e. g., Echeverri et al., 2019; Guo et al., 2022), making pulse field gel electrophoresis separation of Nannochloropsis chromosomes including artificial ones feasible. Such detection method is even easier than FISH that is widely used to visualize the animal and plant artificial chromosomes (Abe et al., 2021). In addition, the reference genome, the parts of artificial chromosomes and all relative elements are known. It is also convenient for us to prove our success in artificial chromosome construction and concatenate gene/DNA fragment insertion by resequencing at a low coverage (Ren et al., 2021). NannoACs loom on the horizon, which will appear in front of us soon (Fig. 1).
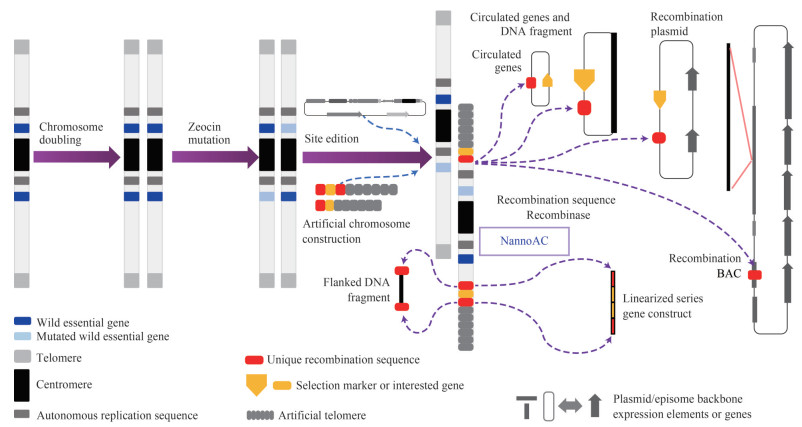
|
| Fig.1 A proposed "top down" construction strategy of Nannochloropsis artificial chromosomes (NannoACs) with a high feasibility |
The molecular tools including genetic transformation, homologous recombination, genome editing, gene stacking and episome vector and diverse reporters and selection markers have been rapidly developing in Nannochloropsis species, the model microalgae for both applications and researches. Unfortunately, the position effect of integrative vectors and the instability of non-integrative ones have hindered genetically engineering Nannochloropsis species. Nannochloropsis artificial chromosomes (NannoACs) are highly appreciated; they should aid to express concatenate genes, stack genes, and transfer very large size DNA fragments which may control a complex trait. Currently, MACs and PACs have set fine examples for NannoACs. It has also been found that two mediate motif-recombinase systems function in Nannochloropsis. The HACs and MACs constructed with "bottom-up" strategy are the circulated and modified centromeres in essence. As the preliminary trials, it is better to truncate natural chromosomes to construct NannoACs (top-down strategy). It seems that the methods and materials (information) to set the foundation for artificial chromosomes in Nannochloropsis species are at hand. A possible and feasible approach to NannoAC construction with top-down strategy has been proposed in this review. NannoACs may contribute the genetic improvement of non-variation Nannochloropsis species and their synthetic biological studies.
6 DATA AVAILABILITY STATEMENTAll data generated and/or analyzed during this study are available from the corresponding author on request.
Abe S, Honma K, Okada A, et al. 2021. Construction of stable mouse artificial chromosome from native mouse chromosome 10 for generation of transchromosomic mice. Scientific Reports, 11(1): 20050.
DOI:10.1038/s41598-021-99535-y |
Abidin A A Z, Suntarajh M, Yusof Z N B. 2020. Transformation of a Malaysian species of Nannochloropsis: gateway to construction of transgenic microalgae as vaccine delivery system to aquatic organisms. Bioengineered, 11(1): 1071-1079.
DOI:10.1080/21655979.2020.1822106 |
Abremski K, Hoess R, Sternberg N. 1983. Studies on the properties of P1 site-specific recombination: evidence for topologically unlinked products following recombination. Cell, 32(4): 1301-1311.
DOI:10.1016/0092-8674(83)90311-2 |
Andersen R A, Brett R W, Potter D, et al. 1998. Phylogeny of the Eustigmatophyceae based upon 18s rDNA, with emphasis on Nannochloropsis. Protist, 149(1): 61-74.
DOI:10.1016/S1434-4610(98)70010-0 |
Araki K, Imaizumi T, Okuyama K, et al. 1997. Efficiency of recombination by Cre transient expression in embryonic stem cells: comparison of various promoters. The Journal of Biochemistry, 122(5): 977-982.
DOI:10.1093/oxfordjournals.jbchem.a021860 |
Arnak R, Bruschi C V, Tosato V. 2012. Yeast artificial chromosomes. In: Encyclopedia of Life Sciences (eLS). John Wiley & Sons Ltd, Chichester, UK, https://doi.org/10.1002/9780470015902.a0000379.pub3.
|
Bailey J C, Freshwater D W. 1997. Molecular systematics of the Gelidiales: inferences from separate and combined analyses of plastid rbcL and nuclear SSU gene sequences. European Journal of Phycology, 32(4): 343-352.
DOI:10.1080/09670269710001737279 |
Boussiba S, Vonshak A, Cohen Z, et al. 1987. Lipid and biomass production by the halotolerant microalga Nannochloropsis salina. Biomass, 12(1): 37-47.
DOI:10.1016/0144-4565(87)90006-0 |
Bruno W J, Knill E, Balding D J, et al. 1995. Efficient pooling designs for library screening. Genomics, 26(1): 21-30.
DOI:10.1016/0888-7543(95)80078-z |
Burke D T, Carle G F, Olson M V. 1987. Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors. Science, 236(4803): 806-812.
DOI:10.1126/science.3033825 |
Carpinelli E C, Telatin A, Vitulo N, et al. 2014. Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion. Molecular Plant, 7(2): 323-335.
DOI:10.1093/mp/sst120 |
Chankova S G, Dimova E, Dimitrova M, et al. 2007. Induction of DNA double-strand breaks by zeocin in Chlamydomonas reinhardtii and the role of increased DNA double-strand breaks rejoining in the formation of an adaptive response. Radiation and Environmental Biophysics, 46(4): 409-416.
DOI:10.1007/s00411-007-0123-2 |
Chen Y W, Hu H H. 2019. High efficiency transformation by electroporation of the freshwater alga Nannochloropsis limnetica. World Journal of Microbiology and Biotechnology, 35(8): 119.
DOI:10.1007/s11274-019-2695-9 |
Chen J W, Huang Y F, Shu Y X, et al. 2022. Recent progress on systems and synthetic biology of diatoms for improving algal productivity. Frontiers in Bioengineering and Biotechnology, 10: 908804.
DOI:10.3389/fbioe.2022.908804 |
Daugbjerg N, Andersen R A. 1997. A molecular phylogeny of the heterokont algae based on analyses of chloroplast-encoded rbcL sequence data. Journal of Phycology, 33(6): 1031-1041.
DOI:10.1111/j.0022-3646.1997.01031.x |
Echeverri D, Romo J, Giraldo N, et al. 2019. Microalgae protoplasts isolation and fusion for biotechnology research. Revista Colombiana de Biotecnología, 21(1): 101-112.
DOI:10.15446/rev.colomb.biote.v21n1.80248 |
Ehrhardt A, Haase R, Schepers A, et al. 2008. Episomal vectors for gene therapy. Current Gene Therapy, 8(3): 147-161.
DOI:10.2174/156652308784746440 |
Elnifro E M, Ashshi A M, Cooper R J, et al. 2000. Multiplex PCR: optimization and application in diagnostic virology. Clinical Microbiology Reviews, 13(4): 559-570.
DOI:10.1128/CMR.13.4.559 |
Fabris M, Abbriano R M, Pernice M, et al. 2020. Emerging technologies in algal biotechnology: toward the establishment of a sustainable, algae-based bioeconomy. Frontiers in Plant Science, 11: 279.
DOI:10.3389/fpls.2020.00279 |
Fawley K P, Fawley M W. 2007. Observations on the diversity and ecology of freshwater Nannochloropsis (Eustigmatophyceae), with descriptions of new taxa. Protist, 158(3): 325-336.
DOI:10.1016/j.protis.2007.03.003 |
Fawley M W, Jameson I, Fawley K P. 2015. The phylogeny of the genus Nannochloropsis (Monodopsidaceae, Eustigmatophyceae), with descriptions of N. australis sp. nov. and Microchloropsis gen. nov.. Phycologia, 54(5): 545-552.
DOI:10.2216/15-60.1 |
Galloway R E. 1990. Selective conditions and isolation of mutants in salt-tolerant, lipid-producing microalgae. Journal of Phycology, 26(4): 752-760.
DOI:10.1111/j.0022-3646.1990.00752.x |
Gee C W, Niyogi K K. 2017. The carbonic anhydrase CAH1 is an essential component of the carbon-concentrating mechanism in Nannochloropsis oceanica. Proceedings of the National Academy of Sciences of the United States of America, 114(17): 4537-4542.
DOI:10.1073/pnas.1700139114 |
Golic K G, Lindquist S. 1989. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell, 59(3): 499-509.
DOI:10.1016/0092-8674(89)90033-0 |
Gong Y H, Kang N K, Kim Y U, et al. 2020. The NanDeSyn database for Nannochloropsis systems and synthetic biology. The Plant Journal, 104(6): 1736-1745.
DOI:10.1111/tpj.15025 |
Gossen M, Bujard H. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proceedings of the National Academy of Sciences of the United States of America, 89(12): 5547-5551.
DOI:10.1073/pnas.89.12.5547 |
Guo C L, Anwar M, Mei R, et al. 2022. Establishment and optimization of PEG-mediated protoplast transformation in the microalga Haematococcus pluvialis. Journal of Applied Phycology, 34(3): 1595-1605.
DOI:10.1007/s10811-022-02718-x |
Guo L, Liang S J, Zhang Z Y, et al. 2019. Genome assembly of Nannochloropsis oceanica provides evidence of host nucleus overthrow by the symbiont nucleus during speciation. Communications Biology, 2(1): 249.
DOI:10.1038/s42003-019-0500-9 |
Halpin C. 2005. Gene stacking in transgenic plants—the challenge for 21st century plant biotechnology. Plant Biotechnology Journal, 3(2): 141-155.
DOI:10.1111/j.1467-7652.2004.00113.x |
Hibberd D J. 1981. Notes on the taxonomy and nomenclature of the algal classes Eustigmatophyceae and Tribophyceae (synonym Xanthophyceae). Botanical Journal of the Linnean Society, 82(2): 93-119.
DOI:10.1111/j.1095-8339.1981.tb00954.x |
Hicks L, Van Der Graaf C M, Childress J, et al. 2018. Streamlined preparation of genomic DNA in agarose plugs for pulsed-field gel electrophoresis. Journal of Biological Methods, 5(1): e86.
DOI:10.14440/jbm.2018.218 |
Ikeno M, Suzuki N. 2011. Construction and use of a bottom-up HAC vector for transgene expression. Methods in Molecular Biology, 738: 101-110.
DOI:10.1007/978-1-61779-099-7_7 |
Kadkhodaei S, Memari H R, Abbasiliasi S, et al. 2016. Multiple overlap extension PCR (MOE-PCR): an effective technical shortcut to high throughput synthetic biology. RSC Advances, 6(71): 66682-66694.
DOI:10.1039/c6ra13172g |
Kandilian R, Lee E, Pilon L. 2013. Radiation and optical properties of Nannochloropsis oculata grown under different irradiances and spectra. Bioresource Technology, 137: 63-73.
DOI:10.1016/j.biortech.2013.03.058 |
Karas B J, Diner R E, Lefebvre S C, et al. 2015. Designer diatom episomes delivered by bacterial conjugation. Nature Communications, 6(1): 6925.
DOI:10.1038/ncomms7925 |
Kilian O, Benemann C S E, Niyogi K K, et al. 2011. High-efficiency homologous recombination in the oil-producing alga Nannochloropsis sp. Proceedings of the National Academy of Sciences of the United States of America, 108(52): 21265-21269.
DOI:10.1073/pnas.1105861108 |
Kouprina N, Petrov N, Molina O, et al. 2018. Human artificial chromosome with regulated centromere: a tool for genome and cancer studies. ACS Synthetic Biology, 7(9): 1974-1989.
DOI:10.1021/acssynbio.8b00230 |
Kurita T, Iwai M, Moroi K, et al. 2022. Genome editing with removable TALEN vectors harboring a yeast centromere and autonomous replication sequence in oleaginous microalga. Scientific Reports, 12(1): 2480.
DOI:10.1038/s41598-022-06495-y |
Kurita T, Moroi K, Iwai M, et al. 2020. Efficient and multiplexable genome editing using Platinum TALENs in oleaginous microalga, Nannochloropsis oceanica NIES-2145. Genes to Cells, 25(10): 695-702.
DOI:10.1111/gtc.12805 |
Kuroiwa Y, Shinohara T, Notsu T, et al. 1998. Efficient modification of a human chromosome by telomere-directed truncation in high homologous recombination-proficient chicken DT40 cells. Nucleic Acids Research, 26(14): 3447-3448.
DOI:10.1093/nar/26.14.3447 |
Kuroiwa Y, Tomizuka K, Shinohara T, et al. 2000. Manipulation of human minichromosomes to carry greater than megabase-sized chromosome inserts. Nature Biotechnology, 18(10): 1086-1090.
DOI:10.1038/80287 |
Le-Feuvre R, Moraga-Suazo P, González-Durán J, et al. 2021. Chemical induction of polyploidy increases astaxanthin accumulation capacity in the microalgae Haematococcus lacustris (Gir. -Chantr.) Rostaf. Algal Research, 59: 102465.
DOI:10.1016/j.algal.2021.102465 |
Lee N C O, Kim J H, Petrov N S, et al. 2018. Method to assemble genomic DNA fragments or genes on human artificial chromosome with regulated kinetochore using a multi-integrase system. ACS Synthetic Biology, 7(1): 63-74.
DOI:10.1021/acssynbio.7b00209 |
Li F J, Gao D W, Hu H H. 2014. High-efficiency nuclear transformation of the oleaginous marine Nannochloropsis species using PCR product. Bioscience, Biotechnology, and Biochemistry, 78(5): 812-817.
DOI:10.1080/09168451.2014.905184 |
Liang C W, Cao S N, Zhang X W, et al. 2013. De novo sequencing and global transcriptome analysis of Nannochloropsis sp. (Eustigmatophyceae) following nitrogen starvation. BioEnergy Research, 6(2): 494-505.
DOI:10.1007/s12155-012-9269-0 |
Lubián L M, Montero O, Moreno-Garrido I, et al. 2000. Nannochloropsis (Eustigmatophyceae) as source of commercially valuable pigments. Journal of Applied Phycology, 12(3): 249-255.
DOI:10.1023/A:1008170915932 |
Ma X N, Chen T P, Yang B, et al. 2016. Lipid production from Nannochloropsis. Marine Drugs, 14(4): 61.
DOI:10.3390/md14040061 |
Manning W M, Strain H H. 1943. Chlorophyll d, a green pigment of red algae. Journal of Biology Chemistry, 151(1): 1-19.
DOI:10.1016/S0021-9258(18)72109-1 |
Mansouri H, Nezhad F S. 2021. Changes in growth and biochemical parameters in Dunaliella salina (Dunaliellaceae) in response to auxin and gibberellin under colchicine-induced polyploidy. Journal of Phycology, 57(4): 1284-1294.
DOI:10.1111/jpy.13173 |
Manzoor A, Ahmad T, Bashir M A, et al. 2019. Studies on colchicine induced chromosome doubling for enhancement of quality traits in ornamental plants. Plants, 8(7): 194.
DOI:10.3390/plants8070194 |
Miga K H, Koren S, Rhie A, et al. 2020. Telomere-to-telomere assembly of a complete human X chromosome. Nature, 585(7823): 79-84.
DOI:10.1038/s41586-020-2547-7 |
Moriwaki T, Abe S, Oshimura M, et al. 2020. Transchromosomic technology for genomically humanized animals. Experimental Cell Research, 390(2): 111914.
DOI:10.1016/j.yexcr.2020.111914 |
Muñoz C F, Südfeld C, Naduthodi M I S, et al. 2021. Genetic engineering of microalgae for enhanced lipid production. Biotechnology Advances, 52: 107836.
DOI:10.1016/j.biotechadv.2021.107836 |
Murata M, Shibata F, Hironaka A, et al. 2013. Generation of an artificial ring chromosome in Arabidopsis by Cre/LoxP-mediated recombination. The Plant Journal, 74(3): 363-371.
DOI:10.1111/tpj.12128 |
Muravenko O, Selyakh I, Kononenko N, et al. 2001. Chromosome numbers and nuclear DNA contents in the red microalgae Cyanidium caldarium and three Galdieria species. European Journal of Phycology, 36(3): 227-232.
DOI:10.1080/09670260110001735378 |
Naduthodi M I S, Claassens N J, D'Adamo S, et al. 2021a. Synthetic biology approaches to enhance microalgal productivity. Trends in Biotechnology, 39(10): 1019-1036.
DOI:10.1016/j.tibtech.2020.12.010 |
Naduthodi M I S, Südfeld C, Avitzigiannis E K, et al. 2021b. Comprehensive genome engineering toolbox for microalgae Nannochloropsis oceanica based on CRISPR-Cas systems. ACS Synthetic Biology, 10(12): 3369-3378.
DOI:10.1021/acssynbio.1c00329 |
Naduthodi M I S, Mohanraju P, Südfeld C, et al. 2019. CRISPR-Cas ribonucleoprotein mediated homology-directed repair for efficient targeted genome editing in microalgae Nannochloropsis oceanica IMET1. Biotechnology for Biofuels, 12(1): 66.
DOI:10.1186/s13068-019-1401-3 |
Nair S, Karim R, Cardosa M J, et al. 1999. Convenient and versatile DNA extraction using agarose plugs for ribotyping of problematic bacterial species. Journal of Microbiological Methods, 38(1-2): 63-67.
DOI:10.1016/s0167-7012(99)00075-5 |
Nezhad F S, Mansouri H. 2019. Induction of polyploidy by colchicine on the green algae Dunaliella salina. Russian Journal of Marine Biology, 45(2): 106-112.
DOI:10.1134/S1063074019020093 |
Oshimura M, Uno N, Kazuki Y, et al. 2015. A pathway from chromosome transfer to engineering resulting in human and mouse artificial chromosomes for a variety of applications to bio-medical challenges. Chromosome Research, 23(1): 111-133.
DOI:10.1007/s10577-014-9459-z |
Pan K H, Qin J J, Li S, et al. 2011. Nuclear monoploidy and asexual propagation of Nannochloropsis oceanica (Eustigmatophyceae) as revealed by its genome sequence. Journal of Phycology, 47(6): 1425-1432.
DOI:10.1111/j.1529-8817.2011.01057.x |
Park S B, Yun J H, Ryu A J, et al. 2021. Development of a novel nannochloropsis strain with enhanced violaxanthin yield for large-scale production. Microbial Cell Factories, 20(1): 43.
DOI:10.1186/s12934-021-01535-0 |
Poliner E, Clark E, Cummings C, et al. 2020. A high-capacity gene stacking toolkit for the oleaginous microalga, Nannochloropsis oceanica CCMP1779. Algal Research, 45: 101664.
DOI:10.1016/j.algal.2019.101664 |
Poliner E, Farré E M, Benning C. 2018a. Advanced genetic tools enable synthetic biology in the oleaginous microalgae Nannochloropsis sp. Plant Cell Reports, 37(10): 1383-1399.
DOI:10.1007/s00299-018-2270-0 |
Poliner E, Pulman J A, Zienkiewicz K, et al. 2018b. A toolkit for Nannochloropsis oceanica CCMP1779 enables gene stacking and genetic engineering of the eicosapentaenoic acid pathway for enhanced long-chain polyunsaturated fatty acid production. Plant Biotechnology Journal, 16(1): 298-309.
DOI:10.1111/pbi.12772 |
Poliner E, Takeuchi T, Du Z Y, et al. 2018c. Nontransgenic marker-free gene disruption by an episomal CRISPR system in the oleaginous microalga, Nannochloropsis oceanica CCMP1779. ACS Synthetic Biology, 7(4): 962-968.
DOI:10.1021/acssynbio.7b00362 |
Ponomartsev S V, Sinenko S A, Skvortsova E V, et al. 2020. Human alphoidtetO artificial chromosome as a gene therapy vector for the developing hemophilia a model in mice. Cells, 9(4): 879.
DOI:10.3390/cells9040879 |
Radakovits R, Jinkerson R E, Fuerstenberg S I, et al. 2012. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropsis gaditana. Nature Communications, 3(1): 686.
DOI:10.1038/ncomms1688 |
Reece-Hoyes J S, Walhout A J M. 2018. Gateway recombinational cloning. Cold Spring Harbor Protocols, 2018(1): pdb.top094912.
DOI:10.1101/pdb.top094912 |
Ren G P, Zhang X, Li Y, et al. 2021. Large-scale whole-genome resequencing unravels the domestication history of Cannabis sativa. Science Advances, 7(29): eabg2286.
DOI:10.1126/sciadv.abg2286 |
Riaz S, Xiao M, Chen P Y, et al. 2021. The genome copy number of the thermophilic cyanobacterium Thermosynechococcus elongatus E542 is controlled by growth phase and nutrient availability. Applied and Environmental Microbiology, 87(9): e02993-20.
DOI:10.1128/AEM.02993-20 |
Ryu A J, Jeong B R, Kang N K, et al. 2021. Safe-harboring based novel genetic toolkit for Nannochloropsis salina CCMP1776: efficient overexpression of transgene via CRISPR/Cas9-mediated knock-in at the transcriptional hotspot. Bioresource Technology, 340: 125676.
DOI:10.1016/j.biortech.2021.125676 |
Sánchez-Gárate J D, Cira-Chavez L A, Rout N P. 2020. Visualization of smaller chromosomes from unicellular microalgae. Brazilian Journal of Botany, 43(3): 633-641.
DOI:10.1007/s40415-020-00619-2 |
Satofuka H, Abe S, Moriwaki T, et al. 2022. Efficient human-like antibody repertoire and hybridoma production in trans-chromosomic mice carrying megabase-sized human immunoglobulin loci. Nature Communications, 13(1): 1841.
DOI:10.1038/s41467-022-29421-2 |
Schweizer H P. 2003. Applications of the Saccharomyces cerevisiae Flp-FRT system in bacterial genetics. Journal of Molecular Microbiology and Biotechnology, 5(2): 67-77.
DOI:10.1159/000069976 |
Sharma-Kuinkel B K, Rude T H, Fowler V G Jr. 2016. Pulse field gel electrophoresis. Methods in Molecular Biology, 1373: 117-130.
DOI:10.1007/7651_2014_191 |
Shizuya H, Kouros-Mehr H. 2001. The development and applications of the bacterial artificial chromosome cloning system. The Keio Journal of Medicine, 50(1): 26-30.
DOI:10.2302/kjm.50.26 |
Südfeld C, Pozo-Rodríguez A, Díez S A M, et al. 2022. The nucleolus as a genomic safe harbor for strong gene expression in Nannochloropsis oceanica. Molecular Plant, 15(2): 340-353.
DOI:10.1016/j.molp.2021.11.003 |
Sukenik A, Carmeli Y, Berner T. 1989. Regulation of fatty acid composition by irradiance level in the eustigmatophyte Nannochloropsis sp. Journal of Phycology, 25(4): 686-692.
DOI:10.1111/j.0022-3646.1989.00686.x |
Suzuki Y, Morishita S. 2021. The time is ripe to investigate human centromeres by long-read sequencing. DNA Research, 28(6): dsab021.
DOI:10.1093/dnares/dsab021 |
Szpyrka E, Broda D, Oklejewicz B, et al. 2020. A non-vector approach to increase lipid levels in the microalga Planktochlorella nurekis. Molecules, 25(2): 270.
DOI:10.3390/molecules25020270 |
Takiguchi M, Kazuki Y, Hiramatsu K, et al. 2014. A novel and stable mouse artificial chromosome vector. ACS Synthetic Biology, 3(12): 903-914.
DOI:10.1021/sb3000723 |
Tan J T, Zhao Y C, Wang B, et al. 2020. Efficient CRISPR/Cas9-based plant genomic fragment deletions by microhomology-mediated end joining. Plant Biotechnology Journal, 18(11): 2161-2163.
DOI:10.1111/pbi.13390 |
Thorpe H M, Smith M C M. 1998. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proceedings of the National Academy of Sciences of the United States of America, 95(10): 5505-5510.
DOI:10.1073/pnas.95.10.5505 |
Tomizuka K, Yoshida H, Uejima H, et al. 1997. Functional expression and germline atransmission of a human chromosome fragment in chimaeric mice. Nature Genetics, 16(2): 133-143.
DOI:10.1038/ng0697-133 |
Torella J P, Boehm C R, Lienert F, et al. 2014. Rapid construction of insulated genetic circuits via synthetic sequence-guided isothermal assembly. Nucleic Acids Research, 42(1): 681-689.
DOI:10.1093/nar/gkt860 |
Verruto J, Francis K, Wang Y J, et al. 2018. Unrestrained markerless trait stacking in Nannochloropsis gaditana through combined genome editing and marker recycling technologies. Proceedings of the National Academy of Sciences of the United States of America, 115(30): E7015-E7022.
DOI:10.1073/pnas.1718193115 |
Vieler A, Wu G X, Tsai C H, et al. 2012. Genome, functional gene annotation, and nuclear transformation of the heterokont oleaginous alga Nannochloropsis oceanica CCMP1779. PLoS Genetics, 8(11): e1003064.
DOI:10.1371/journal.pgen.1003064 |
Wang D M, Ning K, Li J, et al. 2014. Nannochloropsis genomes reveal evolution of microalgal oleaginous traits. PLoS Genetics, 10(1): e1004094.
DOI:10.1371/journal.pgen.1004094 |
Wang Q T, Gong Y H, He Y H, et al. 2021. Genome engineering of Nannochloropsis with hundred-kilobase fragment deletions by Cas9 cleavages. The Plant Journal, 106(4): 1148-1162.
DOI:10.1111/tpj.15227 |
Wang Q T, Lu Y D, Xin Y, et al. 2016. Genome editing of model oleaginous microalgae Nannochloropsis spp. by CRISPR/Cas9. The Plant Journal, 88(6): 1071-1081.
DOI:10.1111/tpj.13307 |
Yang G P, Zhang Z Y, Liu H, et al. 2019. An investigation of the possible methods and potential benefits of de novo cloning of Nannochloropsis oceanica genes. Marine Life Science & Technology, 1(1): 22-27.
DOI:10.1007/s42995-019-00014-1 |
Yu W C, Lamb J C, Han F P, et al. 2006. Telomere-mediated chromosomal truncation in maize. Proceedings of the National Academy of Sciences of the United States of America, 103(46): 17331-17336.
DOI:10.1073/pnas.0605750103 |
Yu W C, Yau Y Y, Birchler J A. 2016. Plant artificial chromosome technology and its potential application in genetic engineering. Plant Biotechnology Journal, 14(5): 1175-1182.
DOI:10.1111/pbi.12466 |
Zerulla K, Ludt K, Soppa J. 2016. The ploidy level of Synechocystis sp. PCC 6803 is highly variable and is influenced by growth phase and by chemical and physical external parameters. Microbiology, 162(5): 730-739.
DOI:10.1099/mic.0.000264 |
Zhang P, Xin Y, He Y H, et al. 2022. Exploring a blue-light-sensing transcription factor to double the peak productivity of oil in Nannochloropsis oceanica. Nature Communications, 13(1): 1664.
DOI:10.1038/s41467-022-29337-x |
Zhang Y, Song W H, Chen S Y, et al. 2021. A bacterial artificial chromosome (BAC)-vectored noninfectious replicon of SARS-CoV-2. Antiviral Research, 185: 104974.
DOI:10.1016/j.antiviral.2020.104974 |
Zhang Y T, Jiang J Y, Shi T Q, et al. 2019. Application of the CRISPR/Cas system for genome editing in microalgae. Applied Microbiology and Biotechnology, 103(8): 3239-3248.
DOI:10.1007/s00253-019-09726-x |
Zulaiha S. 2021. Genetic engineering of microalgae lipid biosynthesis for sustainable biodiesel production. World Journal of Advanced Research and Reviews, 11(3): 72-77.
DOI:10.30574/wjarr.2021.11.3.0397 |
 2023, Vol. 41
2023, Vol. 41


